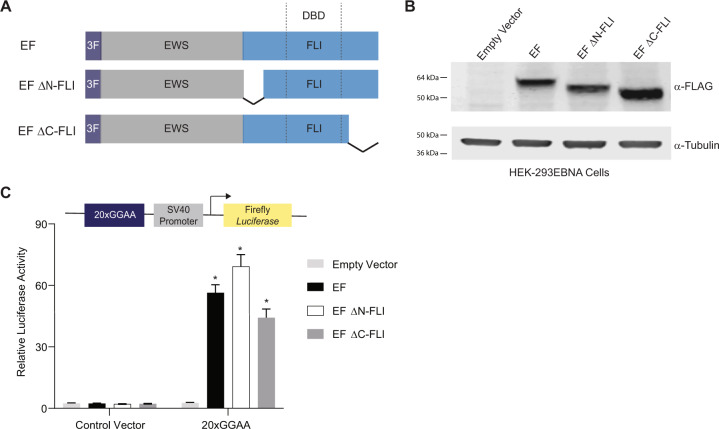
Fig. 1. Amino- and carboxyl-terminal regions of FLI are not required for EWS/FLI-mediated transcriptional activation.
A Protein schematic of 3xFLAG-tagged (3 F) EWS/FLI (EF) cDNA constructs. EWS is represented in gray, FLI is represented in blue, and dashed lines in the FLI portion represent the 85-amino acid ETS DNA-binding domain (DBD) of FLI. In each construct, EWS is fused directly to the FLI portion, but connecting lines are shown here to represent regions of FLI that are eliminated in each construct. EF represents a full-length “type IV” EWS/FLI translocation. EF ΔN-FLI and EF ΔC-FLI indicate constructs where EWS was fused to a version of FLI with a deletion in the amino- or carboxyl-terminal region, respectively. B Western blot of 3xFLAG-tagged EWS/FLI protein expression in HEK-293EBNA cells. Membranes were probed with either α-FLAG or α-tubulin antibodies. C Dual luciferase reporter assay results for the indicated cDNA constructs co-transfected into HEK-293EBNA cells with a Control Vector harboring no GGAA-repeats, or a vector containing 20xGGAA-repeats (represented above the graph). Data are presented as mean ± SEM (N = 6 biological replicates with 3 technical replicates each). Asterisks indicate that the activity of EF, EF ΔN-FLI, and EF ΔC-FLI are each statistically significant when compared to Empty Vector on a 20xGGAA μSat (p-value < 0.05). The activity of EF ΔN-FLI and EF ΔC-FLI are not statistically different from EF on the 20xGGAA μSat (p-value = 0.8).