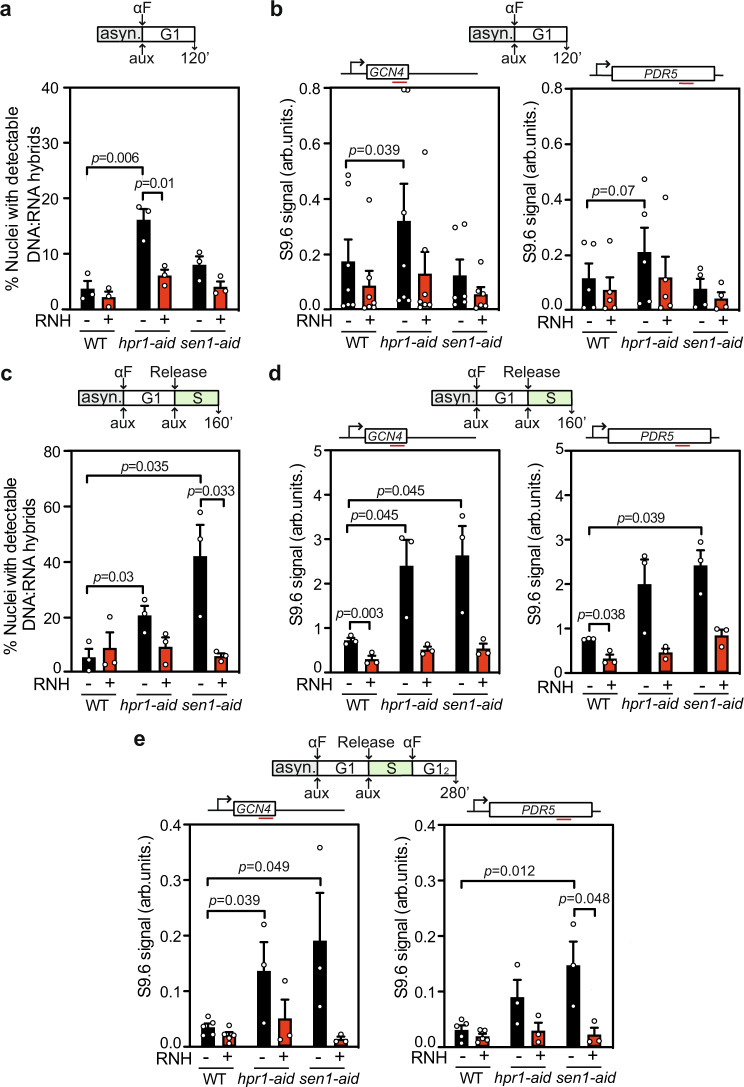
Fig. 3. Hpr1 or Sen1 depletion causes nuclear DNA−RNA hybrid accumulation differently through the cell cycle.
a Percentage of positive nuclei for DNA−RNA hybrids in chromosome spreads stained with the S9.6 antibody in G1-phase WT, hpr1-aid, and sen1-aid cells with or without RNH1. Data are presented as mean values +/− SEM (n = 100 cells examined over three independent experiments). b DRIP with S9.6 antibody at GCN4 and PDR5 genes in G1-phase WT, hpr1-aid, and sen1-aid cells treated (+) or not (−) with RNH1 in vitro. Data are presented as mean values +/− SEM (n = 6 and n = 4 biologically independent experiments for GCN4 and PDR5, respectively). c Percentage of positive nuclei for DNA−RNA hybrids in chromosome spreads stained with the S9.6 antibody in S-phase cells as in (a). Data are presented as mean values +/− SEM (n = 100 cells examined over three independent experiments). d DRIP with S9.6 antibody at GCN4 and PDR5 genes in S-phase cells as in (b), treated (+) or not (−) with RNH1 in vitro. Data are presented as mean values +/− SEM (n = 3 biologically independent experiments). e DRIP with S9.6 antibody at GCN4 and PDR5 genes in G1-phase after one complete cell cycle (G12) cells as in (b), treated (+) or not (−) with RNH1 in vitro. Data are presented as mean values +/− SEM (n = 3 biologically independent experiments). The P values were calculated by the two-tailed unpaired Student t-test. A diagram of the experiment is represented above each plot.