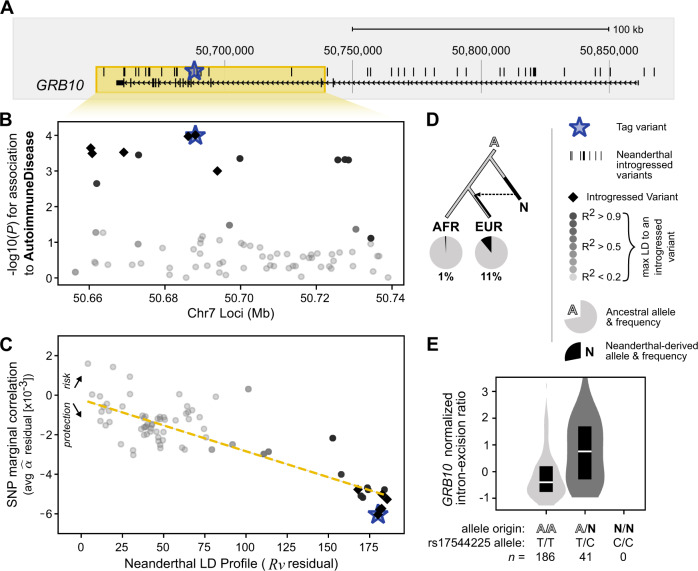
Fig. 4. Signed LD profile regression identifies a candidate functional association between an introgressed haplotype in GRB10 and autoimmune disease.
A A genomic region overlapping GRB10 (chr7: 50,649,920−50,739,129, yellow box) contains an introgressed haplotype. B GWAS Manhattan plot for this region showing associations with autoimmune disease from the UK Biobank (n = 459,324). The strongest single-variant association is at an introgressed variant, but it does not reach genome-wide significance (blue star, P = 9.8 × 10−5 at rs17544225). C Using SLDP regression, we discover a strong negative relationship between LD to introgressed alleles and autoimmune disease in this region. The negative correlation between Neanderthal LD profile and autoimmune disease risk suggests a protective relationship between Neanderthal introgression at this locus and autoimmune disease (r = −0.84). Thus, while the single-variant association alone is not sufficient to implicate this introgressed haplotype in autoimmune disease risk, considering LD to Neanderthal alleles and the direction of effect across variants identifies it as a candidate. D The haplotype (tagged by starred variant rs17544225) is derived in Neanderthals (N) and at 11% frequency in modern Europeans (EUR, n = 503) with 1% frequency in Africans (AFR, n = 661, only observed in admixed African Americans and Caribbeans) (1000 G super-populations). E The introgressed allele is also is an sQTL in which Neanderthal alleles associate with increased GRB10 intron excision in the spleen (two-tailed t-test P = 3 × 10−9). The boxplot centers represent medians and the boxes are bounded by the first and third quartiles.