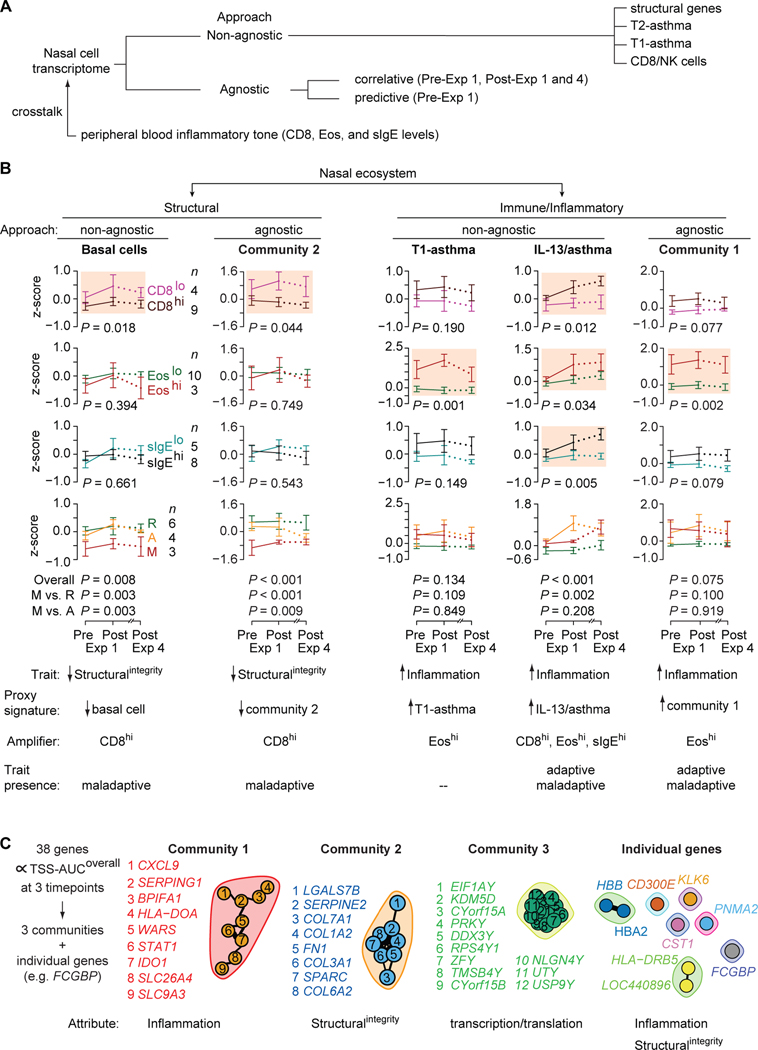
Figure 6. Non-agnostic and agnostic approaches to evaluate nasal cell transcriptomes of persons with HDM-PARC.
(A) Non-agnostic vs. agnostic approaches used to study the nasal cell transcriptomes obtained from HDM-PARC persons. (B) Trajectories of (left to right) structural [basal cells, Community 2] and immune/inflammatory [T1-asthma, IL-13/asthma, Community 1] gene signature scores (z-scores) in nasal cell transcriptomes of HDM-PARC participants at pre- and post-exposure (Exp) 1 and post-Exp 4 stratified by CD8hi vs. CD8lo, Eoshi vs. Eoslo, sIgEhi vs. sIgElo, and HDM-PARC phenotypes (resilient [R], adaptive [A], maladaptive [M]). P, for the difference overall or between the indicated groups. Shaded plots highlight significant differences in signature scores by the indicated peripheral blood traits. Hi, higher; Lo, lower. (C) (Left) 38 correlative genes that, in the nasal cell transcriptomes associated with TSS-AUCoverall at pre- and post-Exp 1 and post-Exp 4. (Right) Depiction of gene communities and single genes. The nodes represent the genes and the edges represent significant correlations (false discovery rate <0.05). Methods for deriving significance values in Supplement.