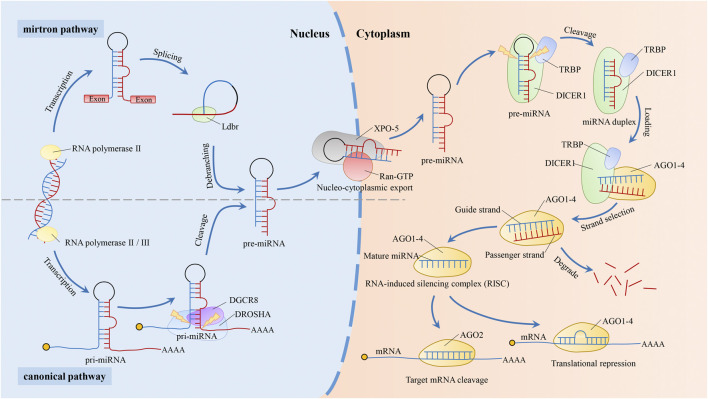
FIGURE 1.
Biogenesis pathway of miRNAs. In the canonical biogenesis pathway of miRNAs. After the miRNA gene is transcribed by RNA polymerase II or III into pri-miRNA, it is sheared by a microprocessor consisting of Drosha and DGCR8 into a pre-miRNA of about 70–100 nucleotides (Rawat et al., 2019). The pre-miRNA is then transported to the cytoplasm by XPO-5 (Yamazawa et al., 2018) and sheared to miRNA duplex by the Dicer-TRBP complex (Fareh et al., 2016; Takahashi et al., 2018). Dicer-TRBP complex binds to AGO1-4 and dissociates after transferring the mature miRNA duplex into AGO1-4 to form RISC. RISC recognizes and binds to target genes by the second to eighth nucleotide counted from 5′-end of the miRNAs (Meijer et al., 2014; Gebert and MacRae, 2019), which is degraded or suppressed the target gene based on the extent of sequence complementarity. Target genes are degraded when completely matched, while inhibited when incompletely matched. During RISC loading, one of the mature miRNAs (the “guide strand”) is retained and forms RISC, while the other is degraded (the “passenger strand”) (Matsuyama and Suzuki, 2019). The non-canonical pathway, mirtron, does not require Drosha. introns are spliced and debranched by the lariat debranching enzyme (Ldbr) to produce pre-miRNA. Similar to the canonical miRNA pathway, this pre-miRNA is transported outside the nucleus via XPO-5. And the mirtron pathway merges with the canonical pathway during this transport stage (Abdelfattah et al., 2014; Stavast and Erkeland, 2019).