Abstract
Background and aims
SARS-CoV-2 has primary pulmonary impairment, but other organs such as the liver can also be affected. This implies a worsening of patient's prognosis and an increase in morbidity and mortality. The metabolic pathways and molecular factors involved in the genesis of this injury are still unknown. Therefore, we aimed to carry out an integrative review about the pathophysiology and possible molecular mechanisms of liver injury by COVID-19.
Methods
We carried out an integrative literature review in the following databases: PubMed, Scopus, and Embase from December 2020 to March 2021 using the following descriptors: # 1 “COVID-19” (MeSH) AND / OR # 2 “Liver injury” (MeSH) AND / OR # 3 “Pathophysiology” (MesH).
Results
The data were extracted and divided into two main themes, for heuristic purposes: “Hepatotropism and SARS-CoV-2”, and “Pathophysiological hypotheses for liver injury associated with SARS-CoV-2”.
Conclusions
The virus seems to promote liver damage through five mechanisms: direct injury, humoral and cellular inflammatory response, hypoxemia caused by a decrease in the effective circulating volume, reinfection through the portal system, and use of drugs in the treatment. The literature also points out that the expression of the angiotensin-converting enzyme II and transmembrane serine protease 2 receptors is expressive in cholangiocyte and is present in hepatocytes, which is a risk factor for the virus to enter these cells. Finally, patients with previous liver disease appear to be more susceptible to liver injury by COVID-19.
Keywords: COVID-19, Liver injury, Pathophysiology, SARS-CoV-2, Gastrointestinal tract
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for the 2019 coronavirus disease (COVID-19), infecting more than 120 million people and causing more than 2.6 million deaths worldwide as of March 15, 2021 [1]. Although SARS-CoV-2 is recognized for causing significant lung damage, extrapulmonary manifestations have been reported around the world [2], including liver damage, both by serum markers and histological evaluation [3].
According to literature data, the correlation between the virus and liver disfunction reaches more than half of the cases in epidemiological studies, which point to the incidence of liver damage associated with cases of COVID-19 infection that range from 4.8% to 53% 3, 4, 5. A meta-analysis that evaluated 15,103 patients with COVID-19 showed that 45.6% of those admitted to hospital units demonstrated a decrease in serum albumin levels, 37.2% showed an increase in aspartate aminotransferase (AST), 26.6% had an alanine aminotransferase (ALT) increase, and 18.2% had raised serum bilirubin levels (95% CI, P < 0,01). During the hospitalization process, the highest incidence found was an increase in AST (69.1%) and a lower incidence of hypoalbuminemia (7.9%) [6].
According to previous studies, the hepatocellular lesion appears to be the result of genetic, environmental, and iatrogenic factors [2,3]. Hematimetric changes, such as leukopenia, neutropenia, and lymphopenia; epidemiological variables, such as male gender and age; use of hepatotoxic medications, such as hydroxychloroquine and tocilizumab; and previous pathologies such as hepatic steatosis are possible markers of worse prognosis in these patients [4,7,8].
However, despite the global collective effort, the factors associated with liver injury have not been well mapped yet. Therefore, we aimed to conduct an integrative literature review in order to understand the molecular and pathophysiological mechanisms of liver injury by COVID-19.
Methods
We carried out an integrative literature review in the following databases: PubMed, Scopus, and Embase. Papers were selected using the following search strategy: # 1 “COVID-19” (MeSH) AND / OR # 2 “Liver injury” (MeSH) AND / OR # 3 “Pathophysiology” (MesH), which was repeated by three independent researchers.
The sample included: papers published between December 2020 and March 2021; manuscripts that addressed a combination of the terms proposed in the search strategy; and papers that discussed the probable direct or indirect pathophysiological mechanisms related to liver injury induced by SARS-CoV-2.
After selection of papers by titles and abstracts, the researchers started reading the texts. The exclusion criteria were: papers repeated in more than one database; papers with fragile methodology or without methodology.
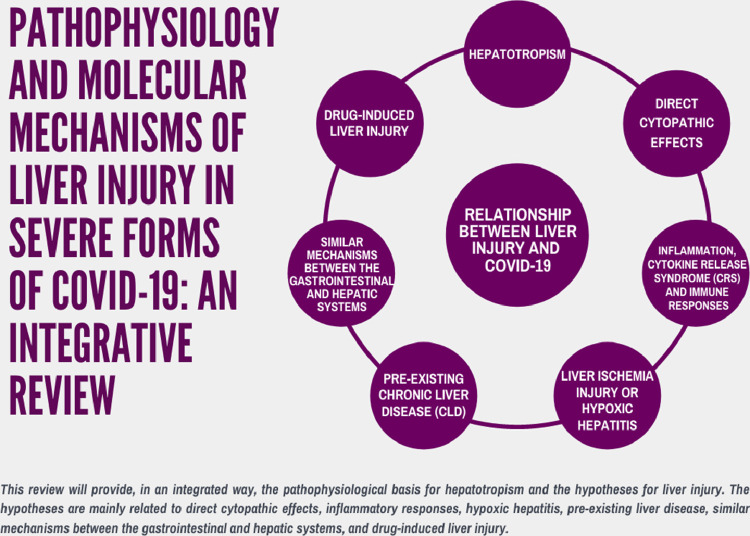
The data were extracted and divided into three main themes for heuristic purposes: hepatotropism and SARS-CoV-2 and pathophysiological hypotheses for liver injury by SARS-CoV-2. Data were also described as mind maps to facilitate the information visualization (Figs. 1 –3). Since this is an integrative literature review, Resolution 510/16 of the Brazilian National Health Council (CNS, acronym in Portuguese) ensures the dispensation of submission to a Human Beings Research Ethics Committee.
Figure 1.
Pathophysiological factors and molecular mechanisms associated with liver injury by COVID-19.
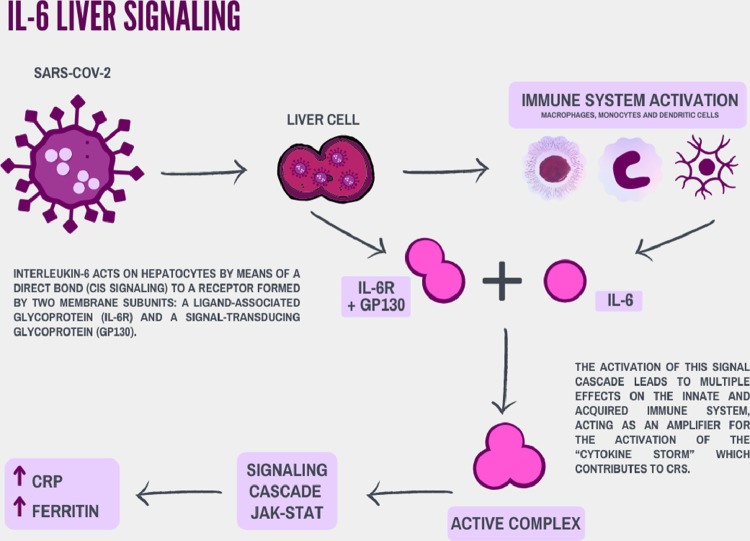
Figure 3.
Participation of IL-6 intracellular signaling in the inflammatory process by SARS-CoV-2 in the hepatocyte.
Discussion
Hepatotropism and SARS-CoV-2
Histopathological analyses have identified the expression of angiotensin-converting enzyme II (ACE2) and transmembrane serine protease 2 (TMPRSS2) receptors in hepatocytes (low expression) and in cholangiocyte (high expression). However, in metabolic stress, up-regulation of these receptors was observed in hepatocytes. These receptors are fundamental for the entry of the virus in human cells and, therefore, for viral hepatotropism [2,3].
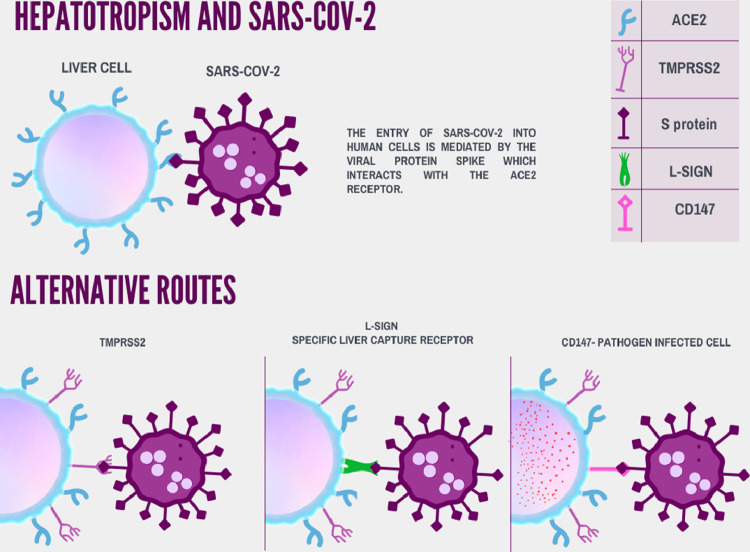
Viral tropism for a specific tissue is determined by the availability of viral receptors on the cell surface [9]. The entry of SARS-CoV-2 into human cells is mediated by the viral protein Spike (S), which interacts with the ACE2 and TMPRSS2 receptors [10] (Figs. 1 and 2 ). There is an expression of ACE2 receptors in hepatocytes at small amounts. In contrast, the incidence of these receptors in the epithelium of the bile duct proved to be similar to that of alveolar cells in the lung [11]. These data are corroborated by experimental studies and computational informatics, in which it is possible to infer the number of ACE2 and TMPRSS2 receptor populations 12, 13, 14, 15 (Figs. 1 and 2).
Figure 2.
Alternative routes and Hepatotropism of the COVID-19 in hepatocytes.
Interestingly, the increase in AST/ALT is greater in patients with COVID-19 compared to gamma-glutamyltransferase (GGT) and alkanine phosphatase (AF). A Chinese study with 156 patients with COVID-19 showed that 41% had AST/ALT elevation 13, 14, 15. The most severe cases are associated with lower levels of albumin, high levels of circulating B and T lymphocytes, higher levels of protein Spikes (S) in the cytoplasm of hepatocytes and dysfunction of organelles such as mitochondria and endoplasmic reticulum [15]. Three possibilities stand out to explain this phenomenon: (I) cell dysfunction due to direct aggression to the virus or/and systemic inflammation; (II) the hepatotoxic potential of the medications used in the treatment for COVID-19 – more explored in the following sessions; (III) other membrane proteins that have not yet been mapped or co-stimulators that influence the binding between ACE2 and S.
In this context, recent study systematically compared differential cell tropism, viral replication kinetics, and cell damage profiles of SARS-CoV-2 and SARS-CoV. The liver cell line Huh7 was the third most susceptible to SARS-CoV-2 replication (p = 0.012), preceded by Calu3 pulmonary cells (p = 0.0003) and Caco2 intestinal cells (p = 0.0009) [13]. Up to 43% of patients with COVID-19 and 44% of patients with SARS developed liver dysfunction. Patients with COVID-19 who needed intensive care had significantly higher amounts of elevated liver necrosis markers. They presented more than three times the upper limit within the normal range when compared with those who did not need intensive care [14,15].
There is also the possibility of alternative pathways to ACE2 receptors for liver infection with SARS-CoV-2 [16]. The liver/lymph node-specific intercellular adhesion molecule-3-grabbing integrin (L-SIGN) receptor is a liver-specific capture receptor for viral infection and immunity [17]. The SARS-CoV glycoprotein can use both ACE2 and L-SIGN in the infection and pathogenesis of the virus [18]. CD147 is another possible receptor for SARS-CoV-2. It is highly expressed in tumor and inflamed tissues, as well as in pathogen-infected cells [19] (Fig. 2). Some indications cite that its binding to protein S may be a new route of entry for SARS-CoV-2 [20].
Pathophysiological hypotheses for liver injury associated with SARS-CoV-2
Direct cytopathic effects
SARS-CoV-2 promotes intracellular cytotoxic actions directly to hepatocytes (Fig. 1), such as destruction of cell membranes and diffuse edema in structures such as rough endoplasmic reticulum and mitochondria. This decreases protein production and compromises hepatocyte ATP biosynthesis [13,21]. However, due to the large population of receptors, especially ACE2 and TMPRSS2 in the cells of the bile ducts, liver damage may start through the bile duct [22]. Alternatively, it is hypothesized that some hepatocytes regenerated after external insults, such as the viral infection itself or previous liver damage, have an important increase in the expression of ACE2 in their membranes, due to compensatory hyperplasia and, therefore, would be more susceptible to reinfection/destruction [23].
Preliminary analyses indicate that SARS-CoV-2 induces decreased mitochondrial activity and oxidative stress in the endoplasmic reticulum 24, 25, 26, 27, 28. At the same time, the virus seems to have the capacity of inducing mitochondrial β-oxidation defects, promoting direct interference in hepatic lipogenesis 24, 25, 26. Therefore, both can be caused by direct cytopathic effects of the new coronavirus, contributing to steatohepatitis secondary to the virus and deteriorating the hepatic metabolic condition, thus aggravating comorbidities such as non-alcoholic fatty liver disease (NAFLD) [10].
The target of rapamycin in mammals (mTOR), an intracytoplasmic enzyme that acts on cell growth, maturation, and proliferation, is also an inducer of lipogenesis and a regulator of autophagy [29,30]. Studies have shown that SARS-CoV-2 restricts autophagy in a manner similar to the mTOR-dependent mechanisms that had already been observed in SARS-CoV and MERS-CoV [31]. In addition, the increase in interleukin-6 (IL-6) promoted by the inflammatory cascade in the virus presence is responsible for activating the mTOR pathway [32]. Hence, direct hepatocyte infection or cytokine storm can stimulate hyperactivation of hepatic mTOR signaling and, therefore, hepatic steatosis in patients affected by COVID-19 [10].
Effects of cytokine release syndrome and immune response
Active replication and virus release by infected cells culminate in the phenomenon of pyroptosis, which is responsible for releasing molecular patterns associated with damage (ATP, nucleic acids). When endothelial cells and alveolar macrophages recognize such cellular products, there is a trigger of proinflammatory cytokines and chemokines, mainly. IL-6, macrophage inflammatory protein 1α (MIP1α), MIP1β and MCP 1 (Fig. 3 ).
The release attracts monocytes, macrophages and T cells to the site, increasing inflammation. The accumulation of immune cells and, therefore, proinflammatory cytokines, constitutes the ‘cytokine storm’ or ‘cytokine release syndrome’ (CRS) (Figs. 1 and 3), damaging the pulmonary structure and afflicting in a multisystemic way to the other organs [33]. IL-6 is involved not only in the acute inflammatory response, but also in liver regeneration and metabolic function of the liver. Molecular signaling of IL-6 occurs through signaling, classical cis or trans signaling pathways. In both signals, IL-6 binds to IL-6R forming a complex with the gp130 dimer. In the classic cis path, the dimerization of the complex formed by IL-6R and gp130 activates the JAK-STAT signaling cascade, contributing to CRS. Hepatocytes can be responsive to IL-6, as some express IL-6R [34,35] (Fig. 3). In the trans signaling pathway, the molecular cascades described before are activated in cells that do not express IL-6R, thus expanding the cell types affected by the ‘cytokine storm’ [36].
In contrast, there is a negative regulation of ACE 2, resulting in the accumulation of angiotensin II (Ang II), in the viral complex endocytosis. Ang II also acts as a proinflammatory cytokine via the AT1R-metalloprotease 17 (ADAM17) axis. ADAM17 can cleave the membrane form of IL-6Ra, thereby generating soluble IL-6R, which binds to IL-6 and subsequently activates STAT signaling. Trans signaling stimulates the production of pro-inflammatory cytokines and chemokines, including IL-6. Therefore, IL-6 can act as an amplifier for CRS activation [23].
The direct correlation between systemic inflammation and liver injuries, mainly indicated by IL-6, C-reactive protein (CRP) and ferritin, has been reported in the literature [37]. IL-6, ferritin and CRP levels were correlated with a significant increase in AST (p < 0.001), in patients hospitalized in and out of the intensive care treatment. Interestingly, higher levels of CRP were found in patients with liver dysfunction without criteria for intensive care. Despite the limitation of a cross-sectional design of this study, it was proposed that the systemic inflammatory response to infection by SARS-CoV-2 in patients with COVID-19 should serve as an incentive for liver injury [38].
Effects of liver ischemic injury or ischemic hepatitis
Changes in body hemodynamics and oxygen supply were also shown to be possible contributors to liver injury [39] (Fig. 1). Experimental studies suggest that reduced oxygen levels and accumulation of lipids in hepatocytes during hypoxemia secondary to CRS can lead to cell death [40]. Thus, the hypoxia-reperfusion injury contributes to liver failure to the extent that it involves a dynamic process of cell injury, which encompasses a dual system comprised of an ischemic phase and an inflammatory response induced by reperfusion [41].
The interruption of adequate blood supply triggers a series of cellular metabolic disorders, leading to a subsequent increase in the reactive oxygen species (ROS) and its peroxidation products [42]. Then, there is activation of transcription factors sensitive to oxidation, amplifying the release of several pro-inflammatory factors (IL-1, IL-6, TNF-alpha) and promoting immune activation of TCD4+ and TCD8+ lymphocytes and macrophages that produce colony stimulating factors (GM-CSF and IFN gamma) in the liver after reperfusion.
Thus, this process involves immune cells of peripheral circulation and several types of non-parenchymal cells, which is a possible generator of hepatocellular lesions [23]. It is understood that in patients with COVID-19 at severe conditions, ischemic hepatitis is an important factor in secondary liver damage. It can be perceived in the laboratory through the marked increase in aminotransferases in the context of respiratory failure, shock, or heart failure [39].
Pre-existing chronic liver disease as a factor of worse prognosis in patients with COVID-19
The effects of pre-existing chronic liver disease (CLD) on the severity of COVID-19 are well documented in the literature as risk factors for more severe forms of COVID-19. A US cohort with 21 centers and 867 patients demonstrated that decompensated cirrhosis, alcohol-reports liver disease (ADL) and hepatocellular carcinoma (HCC) were isolated factors associated with high mortality rates in individuals with CDL [43]. Similar findings were found in the study by Hashimi et al (2020) [44] in which CDL was an isolated risk factor for increasing mortality. Including, data survey carried out between March and April 2020 demonstrates higher mortality by COVID-19 in patients with greater Child-Pugh class. The Child-Pugh class C is up to 3 times more likely to have worse outcomes (death) when compared to the Child-Pugh class A [45]. The US cohort carried out in August 2020 also evaluated 2,780 patients with and without liver disease. The patient with CDL presents higher risk of hospitalization and 4.6 relative risk (RR) for death, compared to controls [46]. However, some early studies lacked this well-documented evidence [47].
Another point to be raised is that in previous studies, CLD is accompanied by other comorbidities such as systemic arterial hypertension, diabetes mellitus, pulmonary disorders, cardiac diseases, drug use such as alcohol and obesity 43, 44, 45, 46. In this context, subgroup analyzes are essential to understand the real impact of CDL in patients with COVID-19. However, this is the clinical reality of most patients.
Really, in theory, due to the systemic immunocompromise state, patients with CLD may be more susceptible to SARS-CoV-2 infection. Patients that present decompensated cirrhosis and comorbidities, such as diabetes and obesity, may have a more severe and progressive acute liver damage [48]. Considering the association with other pre-existing comorbidities, early isolation, intensive surveillance, and timely diagnosis become essential for these patients [23].
Similar mechanisms between gastrointestinal and hepatic systems
Liver and gastrointestinal manifestations have appeared more frequently in the severe forms of COVID-19 infections [49]. A recent study found that patients with gastrointestinal symptoms are more likely to have liver damage than those without these symptoms [50].
Due the various possible clinical scenarios in the context of liver disease, the hypothesis that viruses could enter the portal circulation to reach the liver has proved to be plausible considering that ACE2 is highly expressed on the brush border of small intestine enterocytes. Assuming there is a rapid viral replication in the intestine, Kupffer's liver cells would attempt to eliminate the virus and initiate an inflammatory response, or mediators of inflammatory bowel diseases would enter the portal system and sinusoidal [39]. This statement is corroborated by a meta-analysis developed by Cheung et al. (2020) [51], in which the viral load of SARS-CoV-2 was tested in about 48% of patients and the nucleocapsid SARS-CoV-2 was detected in the cytoplasm of intestinal biopsies, even with negative respiratory samples. On the other hand, Nardo et al. (2020) [10] considered that the gastrointestinal tract may be a primary site of COVID-19 infection and SARS-CoV-2 infection may spread through the hepatobiliary system since the biliary tract provides a direct link between the liver and the intestine. Thus, SARS-CoV-2 could reach and infect the intestine through bile, causing, in turn, a second wave of infection.
Another possibility suggested is the rupture of the intestinal barrier or dysbiosis taking SARS-CoV-2 to the systemic and Portal circulations. Some authors argue that “gut–liver axis disruption” would increase antigenic translocation and activate the immune system [52]. This would occur due to factors such as: (I) local endothelial injury leading to the destruction of tight junctions (TJ) by TNF alpha increase [53]; (II) decrease in native microbiota and increase in opportunistic bacteria (eg.: Rothia spp., Streptococcus spp., Actinomyces spp and Veillonella spp.) in patients with COVID-19 compared to control groups [54]; (III) alteration of ACE2 expression in enterocytes secondary to dysbiosis [55]; and (IV) the presence of the “gut-lung axis”, in which the inflammatory process of COVID-19 would cause mucosal ischemia and bacterial translocation. While translocation would feed back the inflammatory process [56].
Therefore, even though the authors have not reached a consensus about the path of the SARS-CoV-2 virus in the portal circulation yet, there is a strong relationship between the gastrointestinal and hepatic systems, especially with regard to the several forms of COVID-19 infection and greater chances of virus survival, with worse overall outcome in patients who manifest liver and intestinal symptoms in SARS-CoV-2 infection [10,51].
Drug‐induced liver injury
Finally, it is noteworthy that the use of drugs for clinical or off-label tests during the COVID-19 pandemic, in addition to self-medication are factors that contribute to liver injury in patients with COVID-19. This type of injury, called drug-induced liver injury (DILI), is an adverse reaction to drugs or other xenobiotics that occurs as a predictable event when an individual is exposed to toxic doses of some compounds or as an unpredictable event with many drugs of common use [57].
Known hepatotoxic medications have already been used on a large scale, such as antipyretics (acetaminophen), antivirals (remdesivir, lopinavir/ritonavir), antibiotics (macrolides), antimalarials (hydroxychloroquine) and immunomodulators (corticosteroids, tocilizumab).
A hepatotoxic potential has already been confirmed in in vitro/in vivo experiments and in their respective registry studies for most of these drugs (e.g., ritonavir or remdesivir) [10,58]. In this context, several mechanisms have been proposed for DILI. Five highlight here: (I) increased intracellular oxidative stress due to damage to intracytoplasmic organelles; (II) apoptosis measured by Fas pathways to TNF alpha; (III) neutrophil-mediated and T-Lymphocyte-mediated liver destruction [59]; (IV) inhibition of the bile salt export pump (BSEP); (V) mitotoxicity and hepatocyte cytolethality [60]; (V) potential effects of COVID-19 in P450 Cytochrome (CYP-450). Some theories suggest that COVID-19 by CRS, due to the treatment used (eg. glucocorticoids or antivirals drugs) and previous use of other medications (eg. such as antihypertensives, statins and oral hypoglycemic agents). This situation would overload CYP-450, compromising protein metabolism and going through direct liver toxicity [61,62]. Nevertheless, others, with hepatotoxic potential that has not been well recognized yet, seem to have reports in literature, such as tocilizumab [63], whose hepatic metabolism and interference in the IL-6 pathway related to liver regeneration is the most likely etiology for its hepatotoxic effect [64].
Importantly, these mechanisms can be agonists in liver destruction in synergism with dysfunction caused by SARS-CoV-2, CRS caused by COVID-19, ischemia due to septic shock and susceptibility caused by membrane receptors (e.g. CD147, L-SING, ACE2, TMPRSS6).
Conclusion
Therefore, the likely pathophysiological mechanisms for SARS-CoV-2 hepatotropism appear to be closely correlated with the susceptibility of the Huh7 lineage hepatocyte and the presence of the membrane proteins CD147 and L-SIGN to the SARS-CoV-2 protein, which amplify viral invasion and cell dysfunction.
Regarding the pathophysiological mechanisms of the hepatocellular injury of SARS-CoV-2, they are mainly related to the direct cytopathic effects of a probable viral invasion; the increase in populations of ACE2 and TMPRSS6 receptors in bile duct cells, leading to bile duct injury as the genesis of liver disease; hyperactivation of intracytoplasmic mTOR signaling; and CRS with high levels of IL-6. In addition, we highlight not only the role of injury caused by reperfusion injury in the context of circulatory system failure that leads to ischemic hepatitis, but also the use of hepatotoxic drugs in the management of hospitalized or non-hospitalized patients. However, the relationship between gastrointestinal and hepatic symptoms is not a consensus.
Finally, other studies should perform multicenter, observational, and animal-based clinical studies in order to understand and to improve the dynamics of the immune system, the host-parasite process and the inflammatory response in COVID-19, besides to learn how it interferes with liver function.
References
- 1.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20(5):533–534. doi: 10.1016/s1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta A., Madhavan M., Sehgal K., Nair N., Mahajan S., Sehrawat T., et al. Extrapulmonary manifestations of COVID-19. Nat Med. 2020;26(7):1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PubMed] [Google Scholar]
- 3.Lagana S., Kudose S., Iuga A., Lee M., Fazlollahi L., Remotti H., et al. Hepatic pathology in patients dying of COVID-19: a series of 40 cases including clinical, histologic, and virologic data. Mod Pathol. 2020;33(11):2147–2155. doi: 10.1038/s41379-020-00649-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larrey, D., & Pageaux, G. (2004). Hepatotoxicity, drug-induced. encyclopedia of gastroenterology, 354-365. https://doi.org/10.1016/b0-12-386860-2/00194-5
- 5.Xu L., Liu J., Lu M., Yang D., Zheng X. Liver injury during highly pathogenic human coronavirus infections. Liver Int. 2020;40(5):998–1004. doi: 10.1111/liv.14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunutsor S., Laukkanen J. Hepatic manifestations and complications of COVID-19: a systematic review and meta-analysis. J Infect. 2020;81(3):e72–e74. doi: 10.1016/j.jinf.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bin Arif T., Khalid S., Siddiqui M., Hussain H., Sohail H. Incidence, patterns, risk factors, and histopathological findings of liver injury in coronavirus disease 2019 (COVID-19): a scoping review. Hong Kong Med J. 2020 doi: 10.12809/hkmj208732. [DOI] [PubMed] [Google Scholar]
- 8.Sonzogni A., Previtali G., Seghezzi M., Grazia Alessio M., Gianatti A., Licini L., et al. Liver histopathology in severe COVID 19 respiratory failure is suggestive of vascular alterations. Liver Int. 2020;40(9):2110–2116. doi: 10.1111/liv.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chappell J., Duong J., Wright B., Dermody T. Identification of carbohydrate-binding domains in the attachment proteins of type 1 and type 3 reoviruses. J Virol. 2000;74(18):8472–8479. doi: 10.1128/jvi.74.18.8472-8479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nardo A., Schneeweiss-Gleixner M., Bakail M., Dixon E., Lax S., Trauner M. Pathophysiological mechanisms of liver injury in COVID-19. Liver Int. 2020;41(1):20–32. doi: 10.1111/liv.14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chai X., Hu L., Zhang Y., Han W., Lu Z., Ke A., et al. 2020. Specific ACE2 expression in cholangiocytes may cause liver damage after 2019-nCoV infection. [DOI] [Google Scholar]
- 12.Pirola C., Sookoian S. SARS-CoV-2 virus and liver expression of host receptors: putative mechanisms of liver involvement in COVID-19. Liver Int. 2020;40(8):2038–2040. doi: 10.1111/liv.14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu H., Chan J., Yuen T., Shuai H., Yuan S., Wang Y., et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1(1):e14–e23. doi: 10.1016/s2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng V., Lau S., Woo P., Yuen K. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20(4):660–694. doi: 10.1128/cmr.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han D., Lohani M., Cho M. Specific asparagine-linked glycosylation sites are critical for DC-SIGN- and L-SIGN-mediated severe acute respiratory syndrome coronavirus entry. J Virol. 2007;81(21):12029–12039. doi: 10.1128/jvi.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner J., Durso R., Arrigale R., Donovan G., Maddon P., Dragic T., Olson W. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc Natl Acad Sci. 2003;100(8):4498–4503. doi: 10.1073/pnas.0831128100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeffers S., Tusell S., Gillim-Ross L., Hemmila E., Achenbach J., Babcock G., et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci. 2004;101(44):15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong L., Edwards C., Zhou L. The biological function and clinical utilization of CD147 in human diseases: a review of the current scientific literature. Int J Mol Sci. 2014;15(10):17411–17441. doi: 10.3390/ijms151017411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulrich H., Pillat M. CD147 as a target for COVID-19 treatment: suggested effects of azithromycin and stem cell engagement. Stem Cell Rev Rep. 2020;16(3):434–440. doi: 10.1007/s12015-020-09976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonzogni A., Previtali G., Seghezzi M., Grazia Alessio M., Gianatti A., Licini L., et al. Liver histopathology in severe COVID 19 respiratory failure is suggestive of vascular alterations. Liver Int. 2020;40(9):2110–2116. doi: 10.1111/liv.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai Q., Huang D., Yu H., Zhu Z., Xia Z., Su Y., et al. COVID-19: abnormal liver function tests. J Hepatol. 2020;73(3):566–574. doi: 10.1016/j.jhep.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li D., Ding X., Xie M., Tian D., Xia L. COVID-19-associated liver injury: from bedside to bench. J Gastroenterol. 2021;56(3):218–230. doi: 10.1007/s00535-021-01760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tandra S., Yeh M., Brunt E., Vuppalanchi R., Cummings O., Ünalp-Arida A., et al. Presence and significance of microvesicular steatosis in nonalcoholic fatty liver disease. J Hepatol. 2011;55(3):654–659. doi: 10.1016/j.jhep.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koliaki C., Szendroedi J., Kaul K., Jelenik T., Nowotny P., Jankowiak F., et al. Adaptation of hepatic mitochondrial function in humans with non-alcoholic fatty liver is lost in steatohepatitis. Cell Metab. 2015;21(5):739–746. doi: 10.1016/j.cmet.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Rutkowski D., Wu J., Back S., Callaghan M., Ferris S., Iqbal J., et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15(6):829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller B., Silverstein A., Flores M., Xiang W., Cao K., Kumagai H., et al. 2020. SARS-CoV-2 induces a unique mitochondrial transcriptome signature. [DOI] [Google Scholar]
- 28.Chan C., Siu K., Chin K., Yuen K., Zheng B., Jin D. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2006;80(18):9279–9287. doi: 10.1128/jvi.00659-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peterson T., Sengupta S., Harris T., Carmack A., Kang S., Balderas E., et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146(3):408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu G., Sabatini D. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol. 2020;21(4):183–203. doi: 10.1038/s41580-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gassen N., Papies J., Bajaj T., Dethloff F., Emanuel J., Weckmann K., et al. 2020. Analysis of SARS-CoV-2-controlled autophagy reveals spermidine, MK-2206, and niclosamide as putative antiviral therapeutics. [DOI] [Google Scholar]
- 32.He M., Shi X., Yang M., Yang T., Li T., Chen J. Mesenchymal stem cells-derived IL-6 activates AMPK/mTOR signaling to inhibit the proliferation of reactive astrocytes induced by hypoxic-ischemic brain damage. Exp Neurol. 2019;311:15–32. doi: 10.1016/j.expneurol.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 33.Tay M., Poh C., Rénia L., MacAry P., Ng L. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kishimoto T. INTERLEUKIN-6: from basic science to medicine—40 years in immunology. Annu Rev Immunol. 2005;23(1):1–21. doi: 10.1146/annurev.immunol.23.021704.115806. [DOI] [PubMed] [Google Scholar]
- 35.Kang S., Tanaka T., Narazaki M., Kishimoto T. Targeting interleukin-6 signaling in clinic. Immunity. 2019;50(4):1007–1023. doi: 10.1016/j.immuni.2019.03.026. [DOI] [PubMed] [Google Scholar]
- 36.Hunter C., Jones S. IL-6 as a keystone cytokine in health and disease. Nat Immunol. 2015;16(5):448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 37.Effenberger M., Grander C., Grabherr F., Griesmacher A., Ploner T., Hartig F., et al. Systemic inflammation as fuel for acute liver injury in COVID-19. Dig Liver Dis. 2021;53(2):158–165. doi: 10.1016/j.dld.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt-Arras D., Rose-John S. IL-6 pathway in the liver: from physiopathology to therapy. J Hepatol. 2016;64(6):1403–1415. doi: 10.1016/j.jhep.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Bertolini A., Peppel I., Bodewes F., Moshage H., Fantin A., Farinati F., et al. Abnormal liver function tests in patients with COVID-19: relevance and potential pathogenesis. Hepatology. 2020;72(5):1864–1872. doi: 10.1002/hep.31480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang L., Wang W., Wang X., Zhao J., Xiao L., Gui W., et al. Creg in hepatocytes ameliorates liver ischemia/reperfusion injury in a TAK1-dependent manner in mice. Hepatology. 2019;69(1):294–313. doi: 10.1002/hep.30203. [DOI] [PubMed] [Google Scholar]
- 41.Peralta C., Jiménez-Castro M., Gracia-Sancho J. Hepatic ischemia and reperfusion injury: effects on the liver sinusoidal milieu. J Hepatol. 2013;59(5):1094–1106. doi: 10.1016/j.jhep.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 42.Feng G., Zheng K., Yan Q., Rios R., Targher G., Byrne C., et al. COVID-19 and liver dysfunction: current insights and emergent therapeutic strategies. J Clin Transl Hepatol. 2020;8(1):1–7. doi: 10.14218/jcth.2020.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D., Adenjii N., Latt N., Kumar S., Blomm P.P., Aby S.S., et al. Predictors of outcomes of COVID-19 in patients with chronic liver disease: US multi-center study. Clin Gastroenterol Hepatol. 2020 doi: 10.1016/j.cgh.2020.09.027. S1542-3565(20)31288-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hashemi N., Viveiros K., Redd W.D., Zhou J.C., McCarty T.R., Bazarbashi A.N., et al. Impact of chronic liver disease on outcomes of hospitalized patients with COVID-19: a multicentre United States experience. Liver Int. 2020;40(10):2515–2521. doi: 10.1111/liv.14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moon A.M., Weeb G.J., Aloman C., Armstrong M.J., Cargill T., Dhanasekaran R., et al. High mortality rates for SARS-CoV-2 infection in patients with pre-existing chronic liver disease and cirrhosis: preliminary results from an international registry. J Hepatol. 2020;73(3):705–708. doi: 10.1016/j.jhep.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh S., Kham A. Clinical characteristics and outcomes of coronavirus disease 2019 among patients with preexisting liver disease in the United States: a multicenter research network study. Gastroenterology. 2020;159(2):768–771.e3. doi: 10.1053/j.gastro.2020.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lippi G., de Oliveira M., Henry B. Chronic liver disease is not associated with severity or mortality in Coronavirus disease 2019 (COVID-19): a pooled analysis. Eur J Gastroenterol Hepatol. 2020;33(1):114–115. doi: 10.1097/meg.0000000000001742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarin S., Choudhury A., Lau G., Zheng M., Ji D., Abd-Elsalam S., et al. Pre-existing liver disease is associated with poor outcome in patients with SARS CoV2 infection; the APCOLIS Study (APASL COVID-19 Liver Injury Spectrum Study) Hepatol Int. 2020;14(5):690–700. doi: 10.1007/s12072-020-10072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cholankeril G., Podboy A., Aivaliotis V., Pham E., Spencer S., Kim D., Ahmed A. Association of digestive symptoms and hospitalization in patients with SARS-CoV-2 infection. Am J Gastroenterol. 2020;115(7):1129–1132. doi: 10.14309/ajg.0000000000000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin X., Lian J., Hu J., Gao J., Zheng L., Zhang Y., et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69(6):1002–1009. doi: 10.1136/gutjnl-2020-320926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheung K., Hung I., Chan P., Lung K., Tso E., Liu R., et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159(1):81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharma L., Riva A. Intestinal barrier function in health and disease—any role of SARS-CoV-2? Microoganisms. 2020;8:1743–1769. doi: 10.3390/microorganisms8111744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watson A.J.M., Hughes K.R. TNF-α-induced intestinal epithelial cell shedding: implications for intestinal barrier function. Ann N Y Acad Sci. 2012;1258:1–8. doi: 10.1111/j.1749-6632.2012.06523.x. [DOI] [PubMed] [Google Scholar]
- 54.Gu S., Chen Y.Y., Wu Z., Chen Y.Y., Gao H., Lv L., Guo F., Zhang X., Luo R., Huang C., et al. Alterations of the gut microbiota in patients with COVID-19 or H1N1 influenza. Clin Infect Dis. 2020;71(10):2669–2678. doi: 10.1093/cid/ciaa709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang T., Chakraborty S., Saha P., Mell B., Cheng X., Yeo J.Y., Mei X., Zhou G., Mandal J., Golonka R., et al. Gnotobiotic rats reveal that gut microbiota regulates colonic mRNA of Ace2, the receptor for SARS-CoV-2 infectivity. Hypertension. 2020;76:E1–E3. doi: 10.1161/HYPERTENSIONAHA.120.15360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giron L.B., Dweep R., Yin X., Wang H., Damra M., Goldman A.R. Plasma markers of disrupted gut permeability in severe COVID-19 patients. Front Immunol. 2021 doi: 10.3389/fimmu.2021.686240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andrade R., Chalasani N., Björnsson E., Suzuki A., Kullak-Ublick G., Watkins P., et al. Drug-induced liver injury. Nat Rev Dis Primers. 2019;5(1) doi: 10.1038/s41572-019-0105-0. [DOI] [PubMed] [Google Scholar]
- 58.Akinci E., Cha M., Lin L., Yeo G., Hamilton M., Donahue C., et al. 2020. Elucidation of remdesivir cytotoxicity pathways through genome-wide CRISPR-Cas9 screening and transcriptomics. [DOI] [Google Scholar]
- 59.Bertolami M.C. Mechanisms of hepatotoxicity. Arq Bras Cardiol. 2005;85(suppl 5):25–27. doi: 10.1590/S0066-782X2005002400007. [DOI] [PubMed] [Google Scholar]
- 60.Norman B.H. Drug Induced Liver Injury (DILI). Mechanisms and medicinal chemistry avoidance/mitigation strategies. J Med Chem. 2020;63(20):11397–11419. doi: 10.1021/acs.jmedchem.0c00524. [DOI] [PubMed] [Google Scholar]
- 61.Deb S., Arrighi S. Potential effects of COVID-19 on cytochrome P450-mediated drug metabolism and disposition in infected patients. Eur J Drug Metab Pharmacokinet. 2021;46(2):185–203. doi: 10.1007/s13318-020-00668-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pawlos A., Niedzielski M., Gorzelak-Pabiś P., Broncel M., Woźniak E. COVID-19: direct and indirect mechanisms of statins. Int J Mol Sci. 2021;22(8):4177. doi: 10.3390/ijms22084177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muhović D., Bojović J., Bulatović A., Vukčević B., Ratković M., Lazović R., Smolović B. First case of drug-induced liver injury associated with the use of tocilizumab in a patient with COVID-19. Liver Int. 2020;40(8):1901–1905. doi: 10.1111/liv.14516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.National Institute of Diabetes and Digestive and Kidney Diseases; Bethesda (MD): 2012. LiverTox: clinical and research information on drug-induced liver injury [Internet]https://www.ncbi.nlm.nih.gov/books/NBK547852/ Available from: [PubMed] [Google Scholar]