Figure S2.
Epithelial diversity and differentiation in the nasopharyngeal mucosa during COVID-19, related to Figure 2
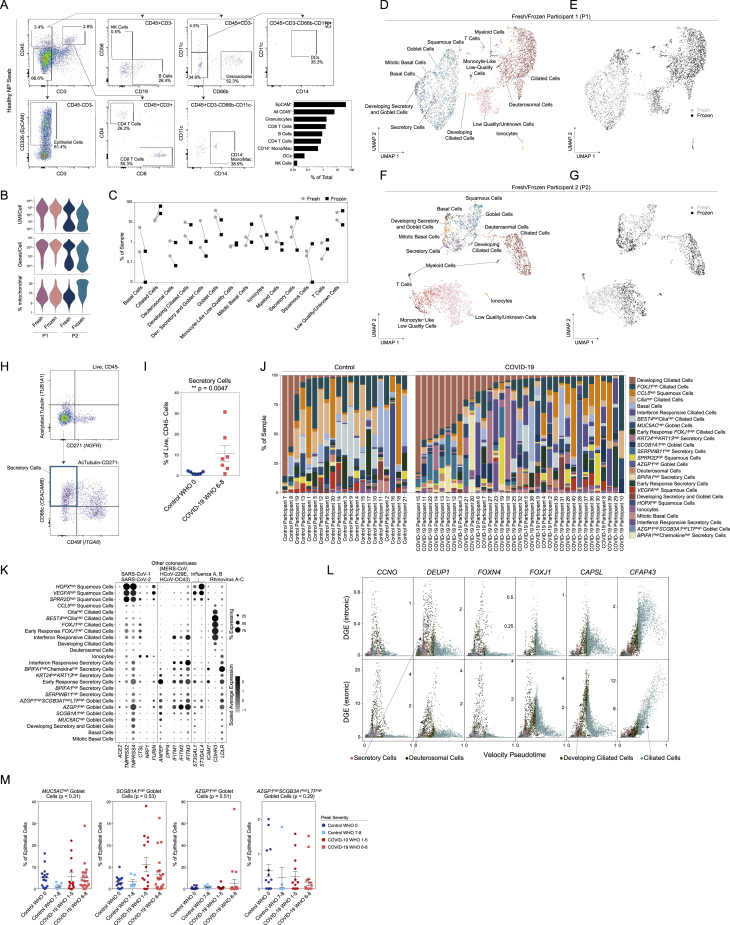
(A) Flow cytometry and gating scheme of immune cells from a fresh nasopharyngeal (NP) swab. Representative healthy participant. Bottom right: quantification of cellular proportions.
(B) Quality metrics for matched fresh versus frozen NP swabs from two healthy participants (P1 and P2).
(C) Percent composition of each cell type by processing type: fresh (gray circles) or frozen (black squares).
(D and E) UMAP of cells from P1, colored by cell types (D) and fresh (gray) versus frozen (black) (E).
(F and G) UMAP of cells from P2, colored by cell types (F) and fresh (gray) versus frozen (black) (G).
(H) Flow cytometry and gating scheme of epithelial cells from an NP swab. Representative data from a participant with severe COVID-19.
(I) Secretory cell proportion of live, CD45- cells from NP swabs. Healthy donors (Control WHO 0): n = 7. Severe COVID-19 (COVID-19 WHO 6-8): n = 7. Secretory cells identified as Live, CD45-ATubulin-CD271-CD49f-CD66c+ cells. Statistical testing: Wilcoxon signed-rank test: ∗∗p = 0.0047.
(J) Proportional abundance of detailed epithelial cell types by participant. Ordered within group by developing ciliated cell proportion.
(K) Expression of entry factors for SARS-CoV-2 and other common upper respiratory viruses among detailed epithelial cell types. Dot size represents fraction of cell type (rows) expressing a given gene (columns). Dot hue represents scaled average expression by gene column.
(L) Plot of gene expression by epithelial cell velocity pseudotime (over all epithelial cells). Select genes significantly associated with ciliated cell pseudotime (FDR < 0.01). Points colored by coarse cell type annotations. Top: alignment to unspliced (intronic) regions. Bottom: alignment to spliced (exonic) regions.
(M) Proportion of goblet cell subtypes (detailed annotation) by sample, normalized to all epithelial cells. Statistical test above graph represents Kruskal-Wallis test results across all groups (following FDR correction).