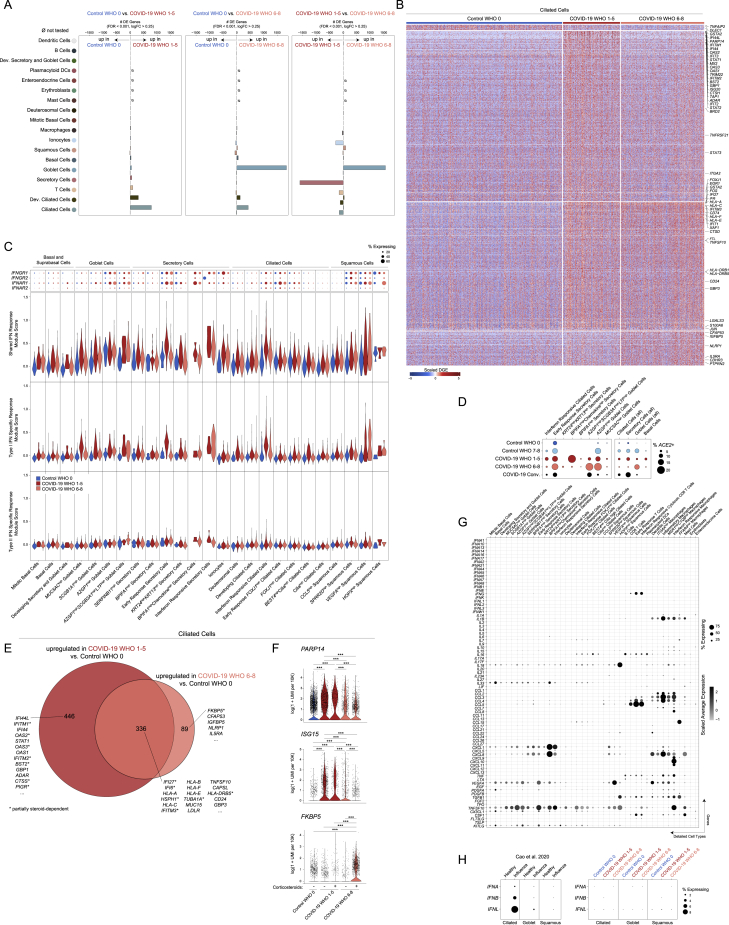
Figure S4.
Cell-type-specific and shared transcriptional Responses to SARS-CoV-2 infection, related to Figure 3
(A) Abundance of significant DE genes by coarse cell type between Control WHO 0 and COVID-19 WHO 1-5 samples (left), Control WHO 0 and COVID-19 WHO 6-8 samples (middle) and COVID-19 WHO 1-5 versus COVID-19 WHO 6-8 samples (right). Gene significance cutoffs: FDR-corrected p < 0.001, log2 fold change > 0.25.
(B) Heatmap of significantly DE genes between ciliated cells (all, coarse annotation) from different disease groups. Values represent row(gene)-scaled digital gene expression (DGE) following log(1+UMI per 10K) normalization.
(C) Top: Dot plot of IFNGR1, IFNGR2, IFNAR1, and IFNAR2 gene expression among all detailed epithelial subtypes. Bottom: Violin plots of module scores, split by Control WHO 0 (blue), COVID-19 WHO 1-5 (red), and COVID-19 WHO 6-8 (pink). Gene modules represent transcriptional responses of human basal cells from the nasal epithelium following in vitro treatment with IFNα or IFNγ. Significance by Wilcoxon signed-rank test. P values following Bonferroni-correction: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(D) Dot plot of ACE2 expression across select epithelial cell types and subsets.
(E) Venn diagram of significantly upregulated genes among ciliated cells between COVID-19 WHO 1-5 versus Control WHO 0 (red) and COVID-19 WHO 6-8 versus Control WHO 0 (pink). Asterisk: genes impacted by corticosteroid treatment within each group.
(F) Violin plots of select genes upregulated among ciliated cells in COVID-19 WHO 1-5 participants compared to Control WHO 0 (PARP14, ISG15) and in COVID-19 WHO 6-8 participants compared to Control WHO 0 (FKBP5). Cells separated by participant treatment with corticosteroids. ∗∗∗ FDR-corrected p < 0.001.
(G) Dot plot of interferon and cytokine expression among detailed epithelial and immune cell types.
(H) Dot plot of type I and type III interferons among ciliated, goblet, and squamous cells. Left: healthy versus influenza A/B virus infected participants from Cao et al., 2020. Right: Control WHO 0 versus COVID-19 WHO 1-5, versus COVID-19 WHO 6-8 participants. Datasets processed and scaled identically.