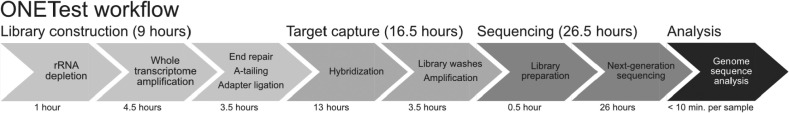
Fig. 1.
The major steps of the ONETest protocol. The ONETest workflow has 4 stages: (1) library construction, (2) target capture, (3) sequencing, and (4) bioinformatics analysis. Input to the ONETest protocol is extracted nucleic acids, or specifically total RNA in this study. Library construction and target capture, which respectively took 9 hours and 16.5 hours, were performed using proprietary kits from Fusion Genomics Corp. Sequencing of the libraries was conducted using an Illumina NextSeq 500 instrument (2 × 150 nt) in this study, and took 26.5 hours. Finally, SARS-CoV-2 genome sequences were reconstructed using the ONETest pipeline described in Materials and Methods, which took less than 10 minutes to run per library. In total, the ONETest workflow in this study took 52 hours.