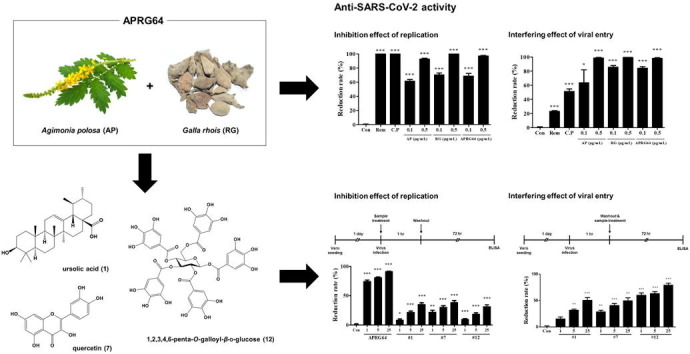
Graphical abstract
Keywords: Antiviral agents, Agrimonia pilosa, APRG64, COVID-19, Galla rhois, SARS-CoV-2
Abbreviations: AP, Agrimonia pilosa; APRG64, 50% EtOH aqueous extract mixture of AP and RG which ratio is 6 and 4; BSA, bovine serum albumin; c.c., column chromatography; COVID-19, Coronavirus disease 2019; DENV-2, dengue virus 2; DMEM, Dulbecco’s Modified Eagle’s Medium; ELISA, Enzyme-linked immunosorbent assay; FBS, fetal bovine serum; HCV, hepatitis C virus; hpi, hours post-infection; HSV-1, herpes simplex virus type 1; IAV, influenza A virus; LC/MS, liquid chromatography/mass spectrometry; MEM, Eagle’s Minimum Essential Medium; ODS, octadecyl SiO2; PFU, plaque-forming unit; RG, Galla rhois; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SEM, standard error of the mean; SiO2, silica gel
Abstract
Agrimonia pilosa (AP), Galla rhois (RG), and their mixture (APRG64) strongly inhibited SARS-CoV-2 by interfering with multiple steps of the viral life cycle including viral entry and replication. Furthermore, among 12 components identified in APRG64, three displayed strong antiviral activity, ursolic acid (1), quercetin (7), and 1,2,3,4,6-penta-O-galloyl-β-d-glucose (12). Molecular docking analysis showed these components to bind potently to the spike receptor-binding-domain (RBD) of the SARS-CoV-2 and its variant B.1.1.7. Taken together, these findings indicate APRG64 as a potent drug candidate to treat SARS-CoV-2 and its variants.
1. Introduction
Since the novel coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in China in December 2019, it has spread rapidly worldwide. The World Health Organization (WHO) declared coronavirus disease 2019 (COVID-19) a global pandemic on March 11, 2020.28 Despite diverse efforts to prevent and control its spread, SARS-CoV-2 is ongoing, and the mortality rate is approximately 2.22% (as of March 5, 202132). Due to the severe threat this poses, development of antiviral drugs targeting the virus is a top priority. Several candidates for the treatment of COVID-19 are being developed. In recent studies, remdesivir and chloroquine effectively suppressed the replication of SARS-CoV-2 in vitro and in pre-clinical studies with murine and nonhuman primate models.31 Although these drug repositioning compounds are being tested clinically for their inhibitory effects on replication of SARS-CoV-2,9 strictly controlled trials have not been completed. Despite the high in vitro antiviral potency of remdesivir against SARS-CoV-2 and some optimistic outcomes in clinical trials,10 uncertainties regarding its side effects (nausea, vomiting, rectal hemorrhage, and hepatic toxicity) and clinical efficacy remain major concerns.11 In addition, another antiviral compound, chloroquine, is not recommended due to safety concerns (side effects on hematologic, hepatic, and renal systems and QTc prolongation with ventricular dysrhythmia).5
Agrimonia pilosa (AP; Rosaceae), commonly known as hairy agrimony, is distributed widely and cultivated in Korea and China, and its aerial parts have been used as an antiviral and for treating hematochezia, traumatic injury, diarrhea, and leukorrhea in Oriental medicine.19, 20, 29 Galla rhois (RG) is the gall caused by the Chinese aphid Schlechtendalia chinensis (Bell) on Rhus chinensis leaves (Anacardiaceae) and has been used in Oriental medicine for treatment of excessive sweating, persistent cough, and diarrhea.34, 4, 6 In our previous work, a 50% EtOH aqueous extract mixture of AP and RG in a 6:4 ratio (APRG64) was optimal to exert maximum biological activities without significant pharmacological toxicity on cardiovascular, central nervous, and respiratory systems.14, 22
Therefore, we hypothesized that AP, RG, and APRG64 will display antiviral activity against SARS-CoV-2. Furthermore, because the emergence of novel variants such as VUI-202012/01 (United Kingdom) and 501.V2 (South Africa) continuously raises concerns for public health, the development of effective antiviral drugs for SARS-CoV-2 and its variants is needed.18, 24 Thus, in the present study, the efficacy of APRG64 for inhibiting the replication of SARS-CoV-2 by interfering with multiple steps of the viral life cycle, including viral entry, was evaluated. In addition, to confirm the possibility as potential therapeutic candidates against novel variant VUI-202012/01, molecular docking of major components of APRG64 was performed at the spike receptor-binding-domain (RBD) of SARS-CoV-2 and its variant B.1.1.7.
2. Materials and methods
2.1. General experimental procedures
The resins used for column chromatography (c.c.) and TLC analysis were described previously.15 Optical rotation, NMR (600 MHz), IR, FABMS, and melting point measurements were performed according to previously reported studies.16 Compounds 2–10 were purchased from Sigma-Aldrich (St. Louis, MO, USA).
2.2. Plant material and isolation of active components
Dried aerial parts of AP and RG were provided by BioKorea Co., Ltd (Seoul, Korea). The plants were identified and authenticated based on macroscopic and microscopic characteristics by Professor Se Chan Kang, Kyung Hee University (Yongin, Korea). Guaranteed samples (BMRI-AP-1601 and BMRI-RG-1602) were deposited in the Laboratory of Natural Medicine Resources in BioMedical Research Institute, Kyung Hee University. The dried aerial parts of AP (500 g) and RG (500 g) were extracted twice with 50% aqueous EtOH (7 L) at room temperature. Each concentrate was divided into EtOAc and H2O fractions. After repeated separation of SiO2 and octadecyl SiO2 (ODS), c.c. of the EtOAc fraction of AP resulted in isolation of ursolic acid (1). In addition, repeated c.c. of the EtOAc fraction of RG resulted in isolation of ethyl gallate (11) and 1,2,3,4,6-penta-O-galloyl-β-d-glucose (12). When comparing their NMR and MS values with those reported in the literature, compounds 1, 11, and 12 were confirmed as ursolic acid,27 ethyl gallate,35 and 1,2,3,4,6-penta-O-galloyl-β-d-glucose,25 respectively (Supplementary Table 1–3 ).
Table 1.
Binding energy calculated from molecular docking analysis of SARS-CoV-2 spike receptor-binding domain (RBD) and B.1.1.7 lineage spike RBD with three anti-SARS-CoV-2 compounds of APRG64, ursolic acid (1), quercetin (7), and 1, 2, 3, 4, 6-penta-O-galloyl-β-d-glucose (12).
| Ligands | Binding energy (kcal/mol) |
|
|---|---|---|
| SARS-CoV-2 spike RBD | B.1.1.7 lineage spike RBD | |
| ursolic acid (1) | −9.5 | −9.0 |
| quercetin (7) | −7.6 | −8.9 |
| 1,2,3,4,6-penta-O-galloyl-β-d-glucose (12) | −3.1 | −3.3 |
Ursolic acid (1): White powder; +70° (c 0.5, CHCl3); m.p. 286–288 °C; IR (KBr, v) 3350, 2900, 1700 cm-1; FABMS m/z 457 [M+H]+; 1H NMR (600 MHz, δ H, pyridine‑d 5) 0.88 (3H, s, H-25), 0.88 (1H, overlapped, H-5), 0.94 (3H, d, J = 6.6 Hz, H-30), 0.98 (1H, overlapped, H-1a), 1.01 (3H, d, J = 6.6 Hz, H-29), 1.01 (3H, s, H-24), 1.03 (1H, overlapped, H-20), 1.05 (3H, s, H-26), 1.19 (1H, m, H-15a), 1.23 (3H, s, H-27), 1.24 (3H, s, H-23), 1.35 (1H, overlapped, H-6a), 1.37 (1H, overlapped, H-7a), 1.37 (1H, overlapped, H-21a), 1.44 (1H, overlapped, H-21b), 1.45 (1H, overlapped, H-19), 1.54 (1H, overlapped, H-1b), 1.56 (1H, overlapped, H-6b),_ 1.57 (1H, overlapped, H-7b), 1.62 (1H, m, H-9), 1.81 (2H, m, H-2), 1.95 (2H, overlapped, H-11), 1.97 (2H, m, H-22), 1.98 (1H, overlapped, H-16a), 2.11 (1H, td, J = 13.8, 3.6 Hz, H-16b), 2.32 (1H, td, J = 13.8, 5.4 Hz, H-15b), 2.63 (1H, d, J = 11.4 Hz, H-18), 3.45 (1H, dd, J = 9.6, 6.0 Hz, H-3), 5.48 (1H, br. s, H-12); 13C NMR (150 MHz, δ C, pyridine‑d 5) 15.5 (C-25), 16.4 (C-24), 17.2 (C-26), 17.3 (C-29), 18.6 (C-6), 21.2 (C-30), 23.4 (C-11), 23.7 (C-27), 24.7 (C-16), 27.9 (C-2), 28.5 (C23), 28.6 (C-15), 30.8 (C-21), 33.4 (C-7), 37.1 (C-22), 37.2 (C-10), 38.8 (C-1), 39.2 (C-20), 39.2 (C-19), 39.3 (C-4), 39.7 (C-8), 42.3 (C-14), 47.8 (C-17), 47.8 (C-9), 53.3 (C-18), 55.6 (C-5), 77.9 (C-3), 125.4 (C-12), 139.1 (C-13), 179.7 (C-28).
Ethyl gallate (11): Colorless needles; m.p. 160–161 °C; IR (KBr, v) 3400, 3300, 2950, 1750 cm-1; FABMS m/z 199 [M+H]+; 1H NMR (600 MHz, δ H, CD3OD) 1.33 (3H, t, J = 7.8 Hz, H-2′), 4.26 (2H, q, J = 7.8 Hz, H-1′), 7.04 (2H, s, H-2,6); 13C NMR (150 MHz, δ C, CD3OD) 14.8 (C-2′), 61.7 (C-1′), 110.0 (C-2,6), 121.8 (C-1), 139.9 (C-4), 148.5 (C-3,5), 168.6 (C-7).
1,2,3,4,6- penta-O-galloyl-β- d -glucose (12): Yellowish amorphous powder; +19° (c 0.5, MeOH); IR (KBr, v) 3400, 1750, 1650 cm-1; FABMS m/z 963 [M+Na]+; 1H NMR (600 MHz, δ H, CD3OD) 4.39 (2H, m, H-6), 4.50 (1H, m, H-5), 5.58 (1H, dd, J = 7.8, 1.8 Hz, H-4), 5.59 (1H, dd, J = 7.8, 7.8 Hz, H-2), 5.89 (1H, dd, J = 7.8, 7.8 Hz, H-3), 6.23 (1H, d, J = 7.8 Hz, H-1), 6.89 (2H, s, H-3-G-2,6), 6.94 (2H, s, H-2-G-2,6), 6.97 (2H, s, H-4-G-2,6), 7.04 (2H, s, H-1-G-2,6), 7.10 (2H, s, H-6-G-2,6); 13C NMR (150 MHz, δ C, CD3OD) 63.1 (C-6), 69.8 (C-4), 72.2 (C-2), 74.1 (C-3), 74.4 (C-5), 93.8 (C-1), 110.3 (C-6-G-2,6), 110.4 (C-3-G-2,6), 110.4 (C-2-G-2,6), 110.5 (C-4-G-2,6), 110.6 (C-1-G-2,6), 119.7 (C-1-G-1), 120.2 (C-2-G-1), 120.4 (C-4-G-1), 120.4 (C-3-G-1), 121.1 (C-6-G-1), 140.0 (C-6-G-3,5), 140.1 (C-3-G-3,5), 140.4 (C-2-G-3,5), 140.4 (C-4-G-3,5), 140.8 (C-1-G-3,5), 146.3 (C-3-G-4), 146.4 (C-2-G-4), 146.5 (C-4-G-4), 146.5 (C-6-G-4), 146.6 (C-1-G-4), 166.2 (C-1-G-7), 166.9 (C-4-G-7), 167.0 (C-2-G-7), 167.3 (C-3-G-7), 168.0 (C-6-G-7).
2.3. Virus, cell, and infection
SARS-CoV-2 (NCCP43326) was provided kindly by the National Culture Collection for Pathogens. Amplification and titration of SARS-CoV-2 were performed on Vero cells as previously described.13, 17 Vero cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with penicillin/streptomycin and 10% fetal bovine serum (FBS). To infect the cells with SARS-CoV-2, Vero cells were incubated for 1 h with the virus in the presence of Eagle’s Minimum Essential Medium (MEM) or DMEM containing 0.3% bovine serum albumin (BSA).
2.4. Plaque reduction assay
The samples were diluted serially from 1/40 to 1/640 with serum-free medium. Vero cells were infected with 50 or 100 plaque-forming units (PFUs) of SARS-CoV-2 and treated with the diluted samples for 1 h at 37 °C in 5% CO2. After washing with PBS twice to remove unbound viruses, the cells were overlaid with 0.6% agarose diluted in DMEM containing 0.3% BSA. The plates were incubated at 37 °C in 5% CO2 for an additional 3 days. The cells were stained with 0.4% crystal violet (Sigma) dye solution for 10 min and rinsed with PBS three times to remove excess dye. The plaque reduction rate in sample-treated cells was calculated by comparison with untreated cells.
2.5. Enzyme-linked immunosorbent assay (ELISA) for SARS-CoV-2 spike proteins
Cell supernatants from virus-infected, sample-treated cells were analyzed for SARS-CoV-2 spike proteins using SARS-CoV-2 spike RBD Nanobody (Cusabio Technology LLC., Houston, TX, USA), according to the manufacturer’s instructions. Briefly, Vero E6 cells were infected with SARS-CoV-2 at 0.3 multiplicity of infection (MOI) and treated with sample or cells were infected with SARS-CoV-2 at 0.3 MOI for 1 h, washed three times with PBS, and then treated with sample. After 72 h, cell supernatants were analyzed for binding activity with SARS-CoV-2 using the SARS-CoV-2 spike RBD Nanobody ELISA kit (Cusabio Technology LLC). The cell supernatant was transferred to an antigen (Recombinant Human Novel Coronavirus Spike glycoprotein)-coated assay plate and incubated for 30 min at 37 °C in 5% CO2. After washing, 100 μL of HRP-conjugate was added to each well and incubated for 30 min at 37 °C in 5% CO2. After washing, 90 μL of TMB substrate was added to each well and incubated for 20 min in the dark at 37 °C in 5% CO2. Then, 50 μL of stop solution was added to each well. The optical density of each well was determined within 10 min using a microplate reader set to 450 nm.
2.6. Statistical analyses
All statistical analyses were performed using Graph Pad Prism 5 software (GraphPad Software Inc., La Jolla, CA, USA). Error bars indicate the standard error of the mean (SEM), and mean values were compared using Student’s t-test. All experiments were repeated independently at least three times.
2.7. Molecular docking of SARS-CoV-2 spike RBD and active components of APRG64
AutoDock (Version 4.2.6, The Scripts Research Institute, La Jolla, CA, USA), an open-source software for molecular docking simulation developed based upon a Lamarckian genetic algorithm (LGA), was utilized to predict the binding affinity between the active components of APRG64 and the SARS-CoV-2 spike receptor-binding domain (RBD). From the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB), the crystal structures of the SARS-CoV-2 spike RBD complexed with its receptor human ACE2 (PDB ID: 6LZG) were retrieved. The three-dimensional (3D) structures of the potent antiviral compounds were downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov) in the structure data file (SDF) format. Components not found in PubChem were modeled using ChemOffice (PerkinElmer, Waltham, MA, USA) to secure the 3D structure data. The ligands were converted to PDB format utilizing Open Babel (Version 2.4.1, http://openbabel.org). Once the SARS-CoV-2 spike RBD and ligands were imported into Autodock, all heteroatoms and water molecules were deleted followed and then polar hydrogens and Kollman charges were introduced before the docking process. The SARS-CoV-2 spike RBD (6LZG) and ligands were converted into PDBQT (Protein Data Bank, Partial Charge (Q), and Atom Type (T)) files. The binding between the SARS-CoV-2 spike RBD and ligands were then examined in Autodock. Blind docking of the ligands was performed to identify the potential primary binding site of the SARS-CoV-2 spike RBD. The entire protein was covered with a grid box dimension of 40 × 62 × 62 Å with a spacing of 1 Å for blind docking and standard LGA was employed. Conformations with high negative binding energy in binding sites were determined and the outcomes were validated through redocking process.
2.8. Molecular docking of B1.1.7 lineage spike RBD and active components of APRG64
The original amino acid sequence of the SARS-CoV-2 spike protein (QHD43416.1) was obtained from GenBank (https://www.ncbi.nlm.nih.gov/genbank). The sites of eight mutations (HV 69-70 deletion, Y144 deletion, N501Y, A570D, P681H, T716I, S982A, and D1118H) were edited specifically to produce the amino acid sequence of the spike protein of a new SARS-CoV-2 lineage called B.1.1.7. The RBD fragment (316–538) were then retrieved from the modified amino acid sequence. The 3D protein structure of the RBD fragment was modeled using SWISS-MODEL (https://swissmodel.expasy.org). This web-based service allows to match the input sequence with similar template sequences in their library (SWISS-MODEL Template Library; SMTL) and model a proper structure in PDB format based on the template structures. In order to validate the structure model, Global Model Quality Estimation (GMQE) and Qualitative Model Energy ANalysis (QMEAN) were evaluated. GMQE is an accuracy index normalized from zero to one while QMEAN is a quality index of which a score below −4.0 indicates poor quality. G-factors and Ramachandran plot were determined to conduct further structure validation using PROCHECK (https://www.ebi.ac.uk/thornton-srv/software/PROCHECK). G-factors are indicatives of structural abnormality of which scores below −0.5 indicate that the given structure is unusual while scores below −1.0 infer that the given structure is highly unusual. Ramachandran plot is a torsional angle (phi and psi) map of the residues, frequently used for validation of a 3D protein structure. A given structure is valid if the residues located in the most favored region in the are greater than 90%. Following structural validation of the B.1.1.7 lineage spike protein, the identical molecular docking protocol was performed to determine the binding energy between the active components of APRG64 and the B.1.1.7 lineage spike RBD and compare with the preceding data with the SARS-CoV-2 spike RBD.
3. Results and discussion
3.1. Inhibition of SARS-CoV-2 replication by AP, RG, and APRG64
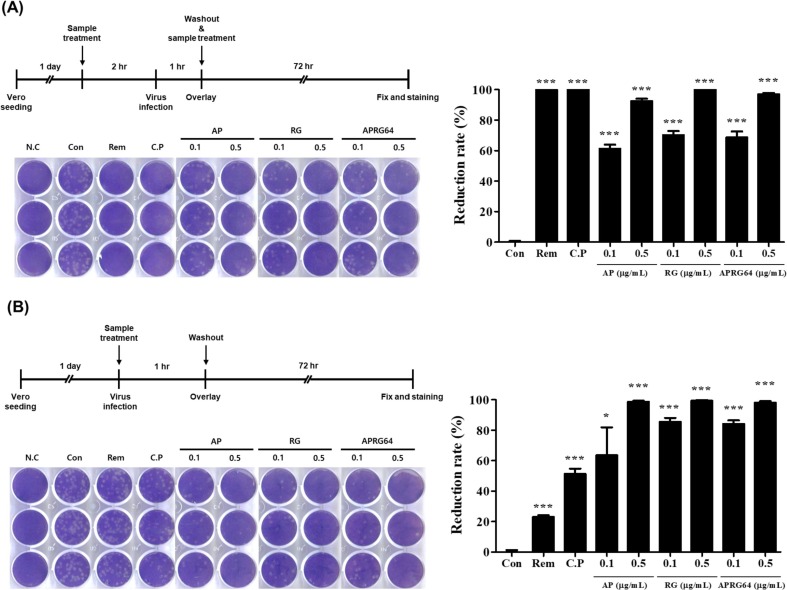
Because AP, RG, and APRG64 have clinical potential for diverse diseases, including diarrhea, cough, chills, and hematochezia, as well as antiviral effects, these extracts were analyzed for antiviral activity against SARS-CoV-2. Vero cells pre-treated with AP, RG, APRG64, remdesivir, or chloroquine phosphate for 2 h were infected with SARS-CoV-2. At 72 h post-infection (hpi), reduction of plaque formations was assessed. Notably, all three extracts strongly inhibited the formation of plaques. Even at low concentrations (0.1 and 0.5 µg/mL), their ability to reduce plaque was comparable to that of remdesivir and chloroquine phosphate (Fig. 1 A). Thus, we conclude that AP, RG, and APRG64 suppressed the replication of SARS-CoV-2.
Fig. 1.
Anti-SARS-CoV-2 activity of Agrimonia pilosa (AP), Galla rhois (RG), and their mixture (APRG64). Vero cells were seeded 1 day before infection. (A) Cells were treated with AP (0.1 or 0.5 µg/mL), RG (0.1 or 0.5 µg/mL), or APRG64 (0.1 or 0.5 µg/mL) for 2 h and then infected with SARS-CoV-2 at 0.01 multiplicity of infection (MOI) for 1 h. After washing three times with PBS, cells were retreated under the same conditions described above. Three days later, cells were fixed and stained to visualize plaques (left panel). Plaque reduction rates are shown in the right panel. (B) Cells were infected with SARS-CoV-2 at 0.01 MOI and treated with AP (0.1 or 0.5 µg/mL), RG (0.1 or 0.5 µg/mL), or APRG64 (0.1 or 0.5 µg/mL). After incubation for 1 h, cells were washed three times with PBS. Three days later, cells were fixed and stained to visualize plaques (left panel). Plaque reduction rates are shown in the right panel. Remdesivir (Rem, 5 µM) and chloroquine phosphate (C.P, 10 µM) were used as positive controls. Data are representative of three independent experiments. *p < 0.05; ***p < 0.001, compared with mock-treated cells (Con).
3.2. Interference with viral entry of SARS-CoV-2 by AP, RG, and APRG64
Next, it was investigated whether AP, RG, and APRG64 influence viral entry, which is a key step for SARS-CoV-2 replication. Vero cells were infected with SARS-CoV-2 in the presence or absence of AP, RG, APRG64, remdesivir, or chloroquine phosphate. After 1 h, cells were washed three times with PBS to remove unattached viruses and extracts from cell culture media. Plaque reduction assay revealed that AP, RG, and APRG64 potently inhibited the replication of SARS-CoV-2. Conversely, remdesivir and chloroquine did not show significant antiviral activity compared with AP, RG, and APRG64 (Fig. 1B). These results strongly support that AP, RG, and APRG64 have potent antiviral activity against SARS-CoV-2 by interfering with viral entry through an antiviral mechanism different from that of remdesivir and chloroquine phosphate.
3.3. Isolation of active components from APRG64
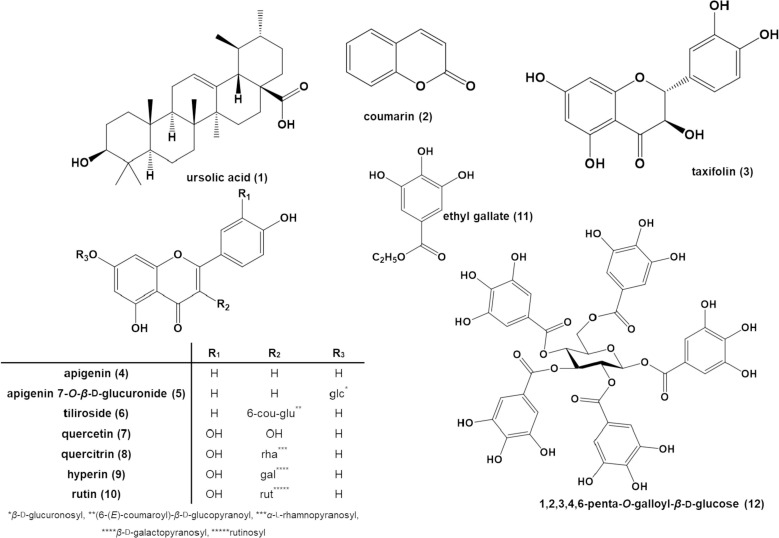
APRG64 is a mixture of AP and RG, and because its safety in vitro as well as in vivo was proven in our previous study,23 its active components were investigated. In repeated c.c. of these plants, ursolic acid (1), ethyl gallate (11), and 1,2,3,4,6-penta-O-galloyl-β-d-glucose (12) were isolated. Based on LC/MS profiling results of APRG64, one triterpenoid (1), one coumarin (2), eight flavonoids (3–10), and two gallate derivatives (11 and 12) were selected as active components of APRG64 and potential anti-COVID-19 agents (Fig. 2 ). These compounds were reported previously as major components of AP1, 8, 36, 17, 7 and RG.23, 21 Compounds 2–10, which were purchased, were analyzed using NMR to evaluate purity. Consequently, purity of all compounds including the isolated compounds (1, 11, and 12) was confirmed to be greater than 97%.
Fig. 2.
Chemical structures of isolated constituents from mixture of Agrimonia pilosa (AP) leaves and Galla rhois (RG) fruits in 50% EtOH extract (APRG64).
3.4. Antiviral activity of APRG64 active components against SARS-CoV-2
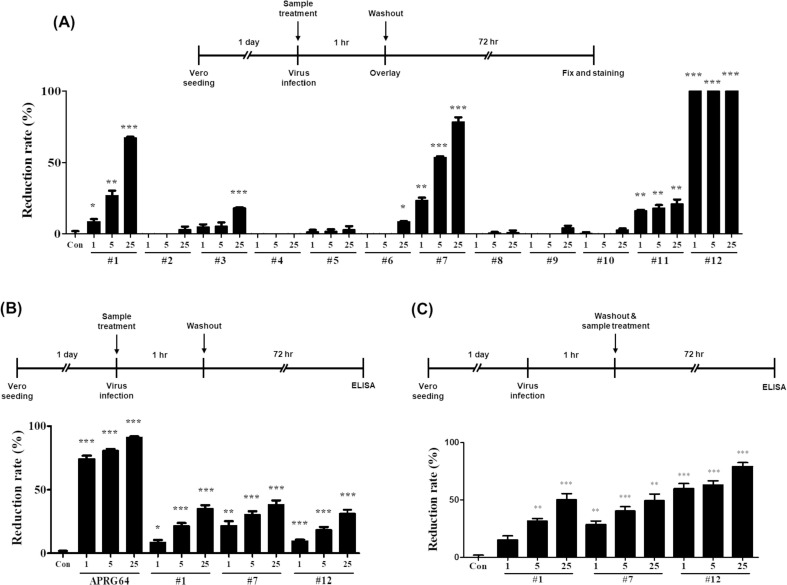
Next, whether secondary metabolites isolated from APRG64 inhibit the replication of SARS-CoV-2 was investigated. Vero cells were infected with SARS-CoV-2 and treated with compounds 1–12, remdesivir, or chloroquine phosphate for 1 h. Cells were washed to eliminate unattached viral particles and compounds. At 72 hpi, the number of plaques was estimated using a plaque assay. As shown in Fig. 3 A, ursolic (1), quercetin (7), ethyl gallate (11), and 1,2,3,4,6-penta-O-galloyl-β-d-glucose (12) significantly inhibited the formation of plaques at all three concentrations (1, 5, and 25 µg/mL). However, ethyl gallate (11) slightly inhibited the formation of plaques (reduction rate: 21.05% at 25 µg/mL), and ursolic acid (1), quercetin (7), and 1,2,3,4,6-penta-O-galloyl-β-d-glucose (12) exhibited much stronger antiviral activity (reduction rates: 78.35%, 67.19%, and 100%, respectively, at 25 µg/mL) (Fig. 3A). In particular, 1,2,3,4,6-penta-O-galloyl-β-d-glucose (12) entirely suppressed the formation of plaques at all three concentrations. Furthermore, 1,2,3,4,6-penta-O-galloyl-β-d-glucose (12) displayed potent antiviral activity at much lower concentrations (0.125, 0.25, 0.5 µg/mL; data not shown). Similar results were obtained when cell supernatants were analyzed for SARS-CoV-2 spike protein. As shown in Fig. 3B, ursolic acid (1), quercetin (7), and 1,2,3,4,6-penta-O-galloyl-β-d-glucose (12) significantly reduced the amount of SARS-CoV-2 spike proteins in cell supernatants (Fig. 3B). Finally, whether post-treatment with ursolic acid (1), quercetin (7), and 1,2,3,4,6-penta-O-galloyl-β-d-glucose (12) was capable of inhibiting SARS-CoV-2 replication was investigated. Vero cells were infected with SARS-CoV-2 for 1 h, followed by three washes to remove unattached viral particles. Cells then were treated with samples for 72 h. Notably, post-treatment with samples significantly reduced SARS-CoV-2 spike proteins in the supernatants (Fig. 3C). Collectively, the results strongly support that active components of APRG64, ursolic acid (1), quercetin (7), and 1,2,3,4,6-penta-O-galloyl-β-d-glucose (12), inhibit SARS-CoV-2 replication by interfering with viral absorption and post-absorption stages. In previous studies, quercetin (7) was reported to have antiviral activities against HIV, herpes simplex virus type 1 (HSV-1),12 hepatitis C virus (HCV),27 influenza A virus (IAV) H1N1, and dengue virus 2 (DENV-2).2, 3, 30 In addition, Yang et al. reported that 1,2,3,4,6-penta-O-galloyl-β-d-glucose (12) has anti-herpes simplex virus activity33; however, the anti-SARS-CoV-2 activity remains unknown.
Fig. 3.
Antiviral activity of active components isolated from APRG64 against SARS-CoV-2. Vero cells were seeded 1 day before infection. (A) Cells were infected with SARS-CoV-2 at 0.01 multiplicity of infection (MOI) and treated with compounds 1–12 of APRG64 at 1, 5, or 25 µg/mL for 1 h, followed by washing three times with PBS. Three days later, cells were fixed and stained to visualize plaques. Plaque reduction rates are shown. (B) Cells were infected with SARS-CoV-2 at 0.3 MOI and treated with APRG64, compounds 1, 7, or 12, at 5 or 25 µg/mL for 1 h. After washing three times with PBS, cells were incubated for an additional 72 h. Cell supernatants were analyzed for the SARS-CoV-2 spike proteins using enzyme-linked immunosorbent assay (ELISA). (C) Cells were infected with SARS-CoV-2 at 0.3 MOI for 1 h, followed by washing three times with PBS. Cells were treated with compounds 1, 7, or 12. After 72 h, cell supernatants were analyzed for SARS-CoV-2 spike protein using ELISA. Data are representative of three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001, compared with mock-treated cells (Con).
3.5. Structural modeling and validation of B.1.1.7 lineage spike RBD
Association between the viral spike RBD and ACE2 proteins on the target cell is a critical step for SARS-CoV-2 to enter the cell. Because APRG64 and its components were shown to interfere with viral absorption (Fig. 1, Fig. 3), the binding of the viral spike RBD onto host cells could be impeded by APRG64 components.
To test this hypothesis, molecular docking analysis of the viral spike RBD onto the structures of three major antiviral components of APRG64, ursolic acid (1), quercetin (7), and 1,2,3,4,6-penta-O-galloyl-β-d-glucose (12), was performed. Notably, the spike RBD of SARS-CoV-2 continuously undergoes mutations, which hinders the development of efficient vaccines and antiviral drugs. Thus, the binding affinity of APRG components to the SARS-CoV-2 spike RBD as well as its variant B.1.1.7 spike RBD was analyzed. First, the B.1.1.7 lineage spike RBD structure model was generated. The spike RBD fragment of the amino acid sequence from the B.1.1.7 lineage was as follows: VQPTESIVRFPNITNLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTYGVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKNKCVNF.
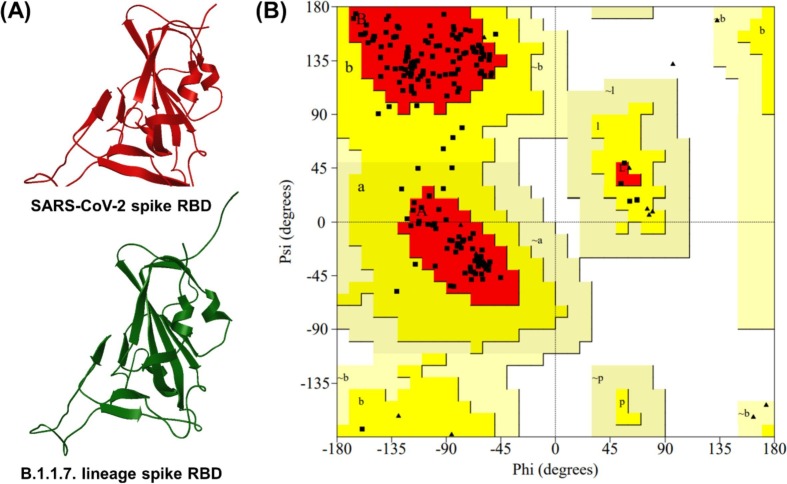
A total of 27 templates was matched to the spike RBD fragment from SMTL and the template 6XC4.1, demonstrating 63% sequence similarity and 99.55% sequence identity compared with the amino acid sequence chosen for structural modeling of the B.1.1.7 lineage spike RBD (Fig. 4 A). The B.1.1.7 lineage spike RBD structure had a total of 199 residues.
Fig. 4.
Structural modeling of B.1.1.7 lineage spike receptor-binding domain (RBD). (A) 3D structures of SARS-CoV-2 spike RBD and B.1.1.7 lineage spike RBD. (B) Ramachandran plot of the B.1.1.7 lineage spike RBD structural model.
For the B.1.1.7 lineage spike RBD structure model, the GMQE and QMEAN values were 0.74 and −1.59, respectively, and the G-factor values obtained from PROCHECK were 0.26 (dihedrals), 0.03 (covalent), and −0.14 (overall). The Ramachandran plot showed 90.6% of the residues to be located in the most favored regions [A, B, L] and 9.4% of the residues in the additionally allowed regions [a, b, l, p] (Fig. 4B). These data indicate that the generated B.1.1.7 lineage spike RBD structure model is stable stereochemically and accurate.
3.6. Comparative molecular docking analysis of SARS-CoV-2 spike RBD and B.1.1.7 lineage spike RBD with active components of APRG64
Comparative molecular docking analyses of the SARS-CoV-2 spike RBD and B.1.1.7 lineage spike RBD with ursolic acid (1), quercetin (7), and 1,2,3,4,6-penta-O-galloyl-β-d-glucose (12) were performed to assess the potential antiviral effects of the active APRG64 components. The binding energy data calculated from the molecular docking analysis are shown in Table 1.
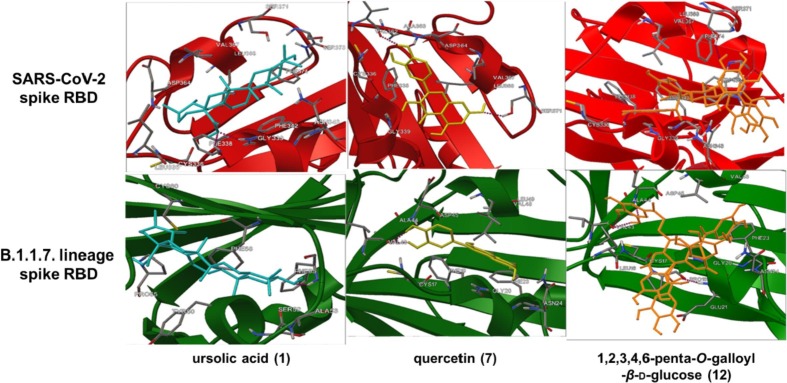
Ursolic acid (1) showed the highest binding energy (−9.5 kcal/mol and −9.0 kcal/mol, respectively) against the SARS-CoV-2 spike RBD and B.1.1.7 lineage spike RBD compared with the other compounds. Notably, quercetin (7) (-8.9 vs. −7.6 kcal/mol) and 1,2,3,4,6-penta-O-galloyl-β-d-glucose (12) (-3.3 vs. −3.1 kcal/mol) showed more stable binding energy when docking with the B.1.1.7 lineage spike RBD compared with docking with the SARS-CoV-2 spike RBD. Fig. 5 represents the hydrogen bond interactions of ursolic acid (1), quercetin (7), and 1,2,3,4,6-penta-O-galloyl-β-d-glucose (12) against the SARS-CoV-2 spike RBD and B.1.1.7 lineage spike RBD. Ursolic acid (1) formed two hydrogen bonds with PHE338 and GLY339 of the SARS-CoV-2 spike RBD amino acid residues and formed one hydrogen bond with PHE55 of the B.1.1.7 lineage spike RBD amino acid residues. These data indicate that APRG64 has significant potential to target both SARS-CoV-2 and B.1.1.7 lineage and exert antiviral effects.
Fig. 5.
Molecular binding of SARS-CoV-2 spike receptor-binding domain (RBD) and B.1.1.7 lineage spike RBD with three active components of APRG64, ursolic acid (1), quercetin (7), and 1, 2, 3, 4, 6-penta-O-galloyl-β-d-glucose (12).
4. Conclusion
In the present study, APRG64 and its active components ursolic acid (1), quercetin (7), and 1,2,3,4,6-penta-O-galloyl-β-d-glucose (12) significantly suppressed the replication of SARS-CoV-2. In addition, the three compounds (1, 7, and 12) showed significant binding affinity for SARS-CoV-2 and its variant (VUI-202012/01). Therefore, APRG64 is a novel candidate antiviral agent to treat COVID-19, and further clinical trials are warranted.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2020R1A6A3A01100042 and 2019R1A2C2010432).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bmc.2021.116329.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.An R.B., Kim H.C., Jeong G.S., Oh S.H., Oh H.C., Kim Y.C. Constituents of the aerial parts of Agrimonia pilosa. Nat Prod Sci. 2005;11:196–198. [Google Scholar]
- 2.Cheng Z., Sun G.e., Guo W., et al. Inhibition of hepatitis B virus replication by quercetin in human hepatoma cell lines. Virol Sin. 2015;30:261–268. doi: 10.1007/s12250-015-3584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiow K.H., Phoon M.C., Putti T., Tan B.K.H., Chow V.T. Evaluation of antiviral activities of Houttuynia cordata Thunb. Extract, quercetin, quercetrin and cinanserin on murine coronavirus and dengue virus infection. Asian Pac J Trop Med. 2016;9:1–7. doi: 10.1016/j.apjtm.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi J.-G., Mun S.-H., Chahar H.S., et al. Methyl gallate from Galla rhois successfully controls clinical isolates of Salmonella infection in both in vitro and in vivo systems. PLoS ONE. 2014;9:e102697. doi: 10.1371/journal.pone.0102697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cortegiani A., Ingoglia G., Ippolito M., Giarratano A., Einav S. A systematic review in the efficacy and safety of chloroquine for the treatment of COVID-19. J Crit Care. 2020;57:279–283. doi: 10.1016/j.jcrc.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djakpo O., Yao W. Rhus chinensis and Galla chinensis–folklore to modern evidence. Phytother Res. 2010;24:1739–1747. doi: 10.1002/ptr.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng J.H., Lee H.J., Kim S.B., Jung J.S., Lim S.S., Suh H.W. Antinociceptive effect of single components isolated from Agrimonia pilosa Ledeb. Extract. Sci Pharm. 2019;87:18. doi: 10.3390/scipharm87030018. [DOI] [Google Scholar]
- 8.Gao Y., Zhou G., Chen J., Qin H. Determination of six flavonoids in Agrimonia pilosa Ledeb by HPLC. Food Sci. 2015;36:93–96. [Google Scholar]
- 9.Grein J., Ohmagari N., Shin D., et al. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holshue M.L., DeBolt C., Lindquist S., et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jean S.-S., Lee P.-I., Hsueh P.-R. Treatment options for COVID-19: The reality and challenges. J Microbiol Immunol Infect. 2020;53:436–443. doi: 10.1016/j.jmii.2020.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaul T.N., Middleton E., Ogra P.L. Antiviral effect of flavonoids on human viruses. J Med Virol. 1985;15:71–79. doi: 10.1002/jmv.1890150110. [DOI] [PubMed] [Google Scholar]
- 13.Kim J.-M., Chung Y.-S., Jo H.J., et al. Identification of coronavirus isolated from a patient in Korea with COVID-19. Osong Public Health Res Perspect. 2020;11:3–7. doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon J.E., Lee Y.-G., Kang J.-H., et al. Anti-viral activity of compounds from Agrimonia pilosa and Galla rhois extract mixture. Bioorg Chem. 2019;93:103320. doi: 10.1016/j.bioorg.2019.103320. [DOI] [PubMed] [Google Scholar]
- 15.Lee Y.-G., Lee H., Jung J.-W., et al. Flavonoids from Chionanthus retusus (Oleaceae) flowers and their protective effects against glutamate-induced cell toxicity in HT22 cells. Int J Mol Sci. 2019;20:3517. doi: 10.3390/ijms20143517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y.-G., Lee H., Ryuk J.A., et al. 6-Methoxyflavonols from the aerial parts of Tetragonia tetragonoides (Pall.) Kuntze and their anti-inflammatory activity. Bioorg Chem. 2019;88:102922. doi: 10.1016/j.bioorg.2019.102922. [DOI] [PubMed] [Google Scholar]
- 17.Liu W.-J., Hou X.-Q., Chen H., Liang J.-y., Sun J.-b. Chemical constituents from Agrimonia pilosa Ledeb. and their chemotaxonomic significance. Nat Prod Res. 2016;30:2495–2499. doi: 10.1080/14786419.2016.1198351. [DOI] [PubMed] [Google Scholar]
- 18.Loconsole D., Sallustio A., Accogli M., et al. Genome sequence of a sars-cov-2 vui 202012/01 strain identified from a patient returning from London, England, to the apulia region of Italy. Microbiol Resour Announc. 2021;10:e01487–e1520. doi: 10.1128/MRA.01487-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Min B.S., Kim Y.H., Tomiyama M., et al. Inhibitory effects of Korean plants on HIV-1 activities. Phytothe Res. 2001;15:481–486. doi: 10.1002/ptr.751. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen D.H., Seo U.M., Zhao B.T., et al. Ellagitannin and flavonoid constituents from Agrimonia pilosa L. with their protein tyrosine phosphatase and acetylcholinesterase inhibitory activities. Bioorg Chem. 2017;72:293–300. doi: 10.1016/j.bioorg.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 21.Park E.J., Zhao Y.Z., An R.B., Kim Y.C., Sohn D.H. 1,2,3,4,6-penta-O-galloyl-β-D-glucose from Galla rhois protects primary rat hepatocytes from necrosis and apoptosis. Planta Med. 2008;74:1380–1383. doi: 10.1055/s-2008-1081300. [DOI] [PubMed] [Google Scholar]
- 22.Park J.-H., Ra J.-S., Kwon J.E., et al. Evaluation of genetic toxicity, acute and sub-chronic oral toxicity and systemic safety of Agrimonia pilosa and Rhus gall 50% ethanolic extract mixture (APRG64) in vitro and in vivo (rodent and non-rodent animal models) Toxicol Res. 2020;36:367–406. doi: 10.1007/s43188-020-00042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park P.-H., Hur J., Kim Y.-C., An R.-B., Sohn D.H. Involvement of heme oxygenase-1 induction in inhibitory effect of ethyl gallate isolated from Galla rhois on nitric oxide production in RAW 264.7 macrophages. Arch Pharm Res. 2011;34:1545–1552. doi: 10.1007/s12272-011-0917-2. [DOI] [PubMed] [Google Scholar]
- 24.Peñarrubia L., Ruiz M., Porco R., et al. In Response to: Multiple assays in a real-time RT-PCR Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) panel can mitigate the risk of loss of sensitivity by new genomic variants during the COVID-19 outbreak. Int J Infect Dis. 2021;105:241–242. doi: 10.1016/j.ijid.2021.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piao X., Piao X.-L., Kim H.Y., Cho E.J. Antioxidative activity of geranium (Pelargonium inquinans Ait) and its active component, 1, 2, 3, 4, 6-penta-O-galloyl-β-D-glucose. Phytother Res. 2008;22:534–538. doi: 10.1002/ptr.2398. [DOI] [PubMed] [Google Scholar]
- 26.Rojas Á., Del Campo J.A., Clement S., et al. Effect of quercetin on hepatitis C virus life cycle: from viral to host targets. Sci Rep. 2016;6 doi: 10.1038/srep31777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seebacher W., Simic N., Weis R., Saf R., Kunert O. Complete assignments of 1H and 13C NMR resonances of oleanolic acid, 18α-oleanolic acid, ursolic acid and their 11-oxo derivatives. Magn Reson Chem. 2003;41:636–638. [Google Scholar]
- 28.Shan C., Yao Y.-F., Yang X.-L., et al. Infection with novel coronavirus (SARS-CoV-2) causes pneumonia in Rhesus macaques. Cell Res. 2020;30:670–677. doi: 10.1038/s41422-020-0364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin W.J., Lee K.H., Park M.H., Seong B.L. Broad-spectrum antiviral effect of Agrimonia pilosa extract on influenza viruses. Microbiol Immunol. 2010;54:11–19. doi: 10.1111/j.1348-0421.2009.00173.x. [DOI] [PubMed] [Google Scholar]
- 30.Vaidya B., Cho S.-Y., Oh K.-S., et al. Effectiveness of periodic treatment of quercetin against influenza a virus H1N1 through modulation of protein expression. J Agric Food Chem. 2016;64:4416–4425. doi: 10.1021/acs.jafc.6b00148. [DOI] [PubMed] [Google Scholar]
- 31.Wang M., Cao R., Zhang L., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.WHO. WHO Coronavirus Disease (COVID-19) Dashboard; 2021.
- 33.Yang C.-M., Cheng H.-Y., Lin T.-C., Chiang L.-C., Lin C.-C. The in vitro activity of geraniin and 1,3,4,6-tetra-O-galloyl-β-D-glucose isolated from Phyllanthus urinaria against herpes simplex virus type 1 and type 2 infection. J Ethnopharmacol. 2007;110:555–558. doi: 10.1016/j.jep.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 34.Yim N.-H., Gu M.J., Hwang Y.-H., Cho W.-K., Ma J.Y. Water extract of Galla rhois with steaming process enhances apoptotic cell death in human colon cancer cells. Integr Med Res. 2016;5:284–292. doi: 10.1016/j.imr.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang W., Li C., You L.-J., Fu X., Chen Y.-S., Luo Y.-Q. Structural identification of compounds from Toona sinensis leaves with antioxidant and anticancer activities. J Funct Foods. 2014;10:427–435. [Google Scholar]
- 36.Zhu L., Chen J., Tan J., Liu X.i., Wang B. Flavonoids from Agrimonia pilosa Ledeb: free radical scavenging and DNA oxidative damage protection activities and analysis of bioactivity-structure relationship based on molecular and electronic structures. Molecules. 2017;22:195. doi: 10.3390/molecules22030195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.