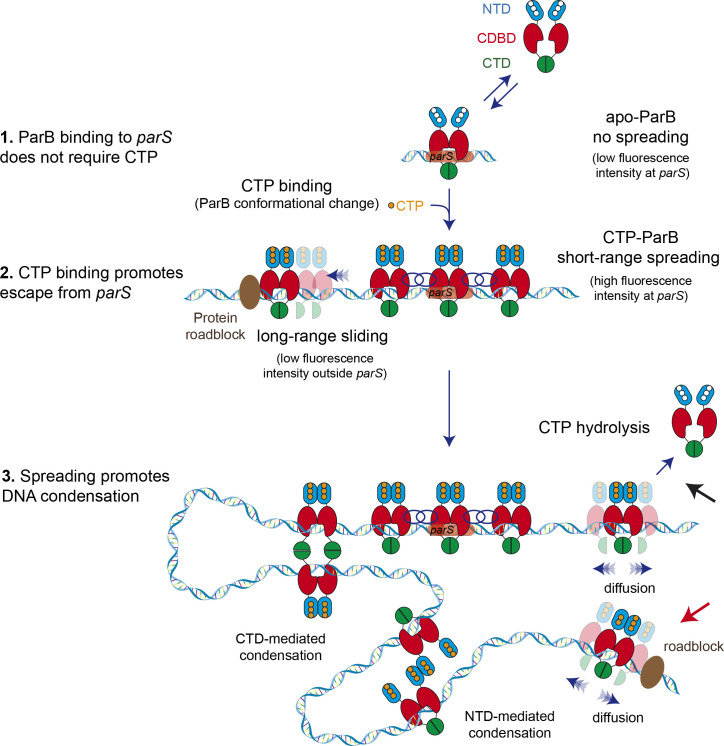
Figure 7. Model for ParB-dependent DNA condensation around parS sequences.
(Step 1) ParB binding to parS does not require cytidine triphosphate (CTP) as observed from C-trap experiments. ParS-bound apo-ParB does not spread from parS. (Step 2) CTP binding to ParB induces a conformational change to a sliding clamp which then escapes from parS to neighbouring non-specific DNA. Potential interactions between the ParB proteins around parS are represented by interlaced blue circles. Some ParB proteins are able to slide/diffuse long distances. (Step 3) ParB spreading and diffusion promotes the interaction with other CTP-ParB dimers through the C-terminal domain (CTD) of ParB (Fisher et al., 2017), resulting in DNA condensation by forming large DNA loops. Alternatively, other protein-protein interaction such those mediated by the N-terminal domain (NTD) (shown in figure) or the central DNA-binding domain (CDBD) of ParB could result in DNA condensation. CTP hydrolysis might be a means to recover ParB dimers from the DNA (black arrow). Protein roadblocks constrain diffusion of ParB proteins (red arrow).