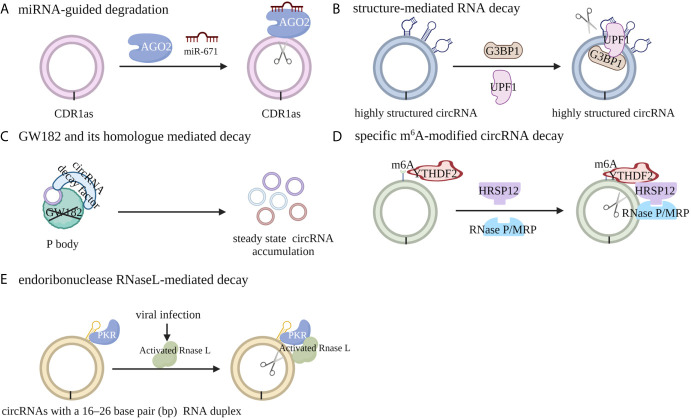
Figure 2.
Mechanisms of circRNA degradation. (A) CircRNA CDR1 can be cleaved by endonuclease AGO2 with the assistance of miR-671 in a miRNA target sequence-dependent manner. (B) The degradation of highly structured circRNAs can be regulated by UPF1 and G3BP1, both of which recognize and unwind the overall structures of circRNAs. (C) m6A-modified circRNA can recruit the m6A reader protein YTHDF2 as well as the adaptor protein HRSP12, and HRSP12 can serve as a bridge to connect YTHDF2 with the endoribonuclease RNase P/MRP, thus enabling degradation of m6A modified circRNA by RNase P/MRP. (D) The absence of GW182 or its human homologue TRNC6A/B/C, the key component of P body and RNAi complexes, can lead to the accumulation of endogenous circRNA and increase the steady state of cytoplasmic circRNAs. (E) Upon viral infection, RNase L can be activated and can degrade circRNAs with RNA duplexes containing 16 to 26 bp through an undefined mechanism.