Extended Data Fig. 5 |. VbP interactions in the DPP9 active site and comparison to a DPP substrate and NLRP1.
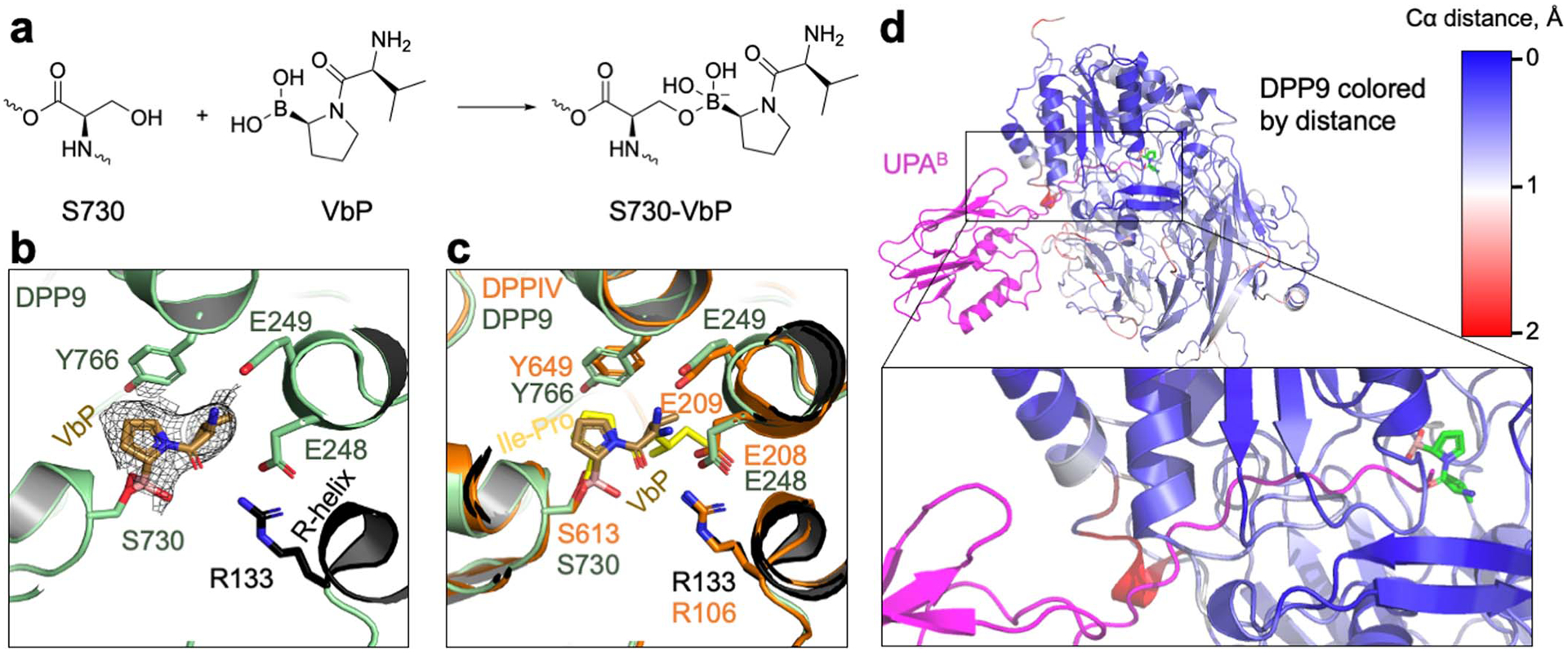
a, Schematic of covalent linkage between DPP9’s S730 and VbP. b, Fit of VbP into the cryo-EM density. VbP is shown in stick with carbon atoms in light brown. The charged amino group of VbP interacts with the DPP9 EE loop which also coordinates a substrate N-terminus, and the carbonyl oxygen of VbP interacts with R133 of the R-helix. The covalent linkage of VbP with S730, the catalytic serine, is displayed. c, Structural alignment of the VbP-bound DPP9 model (green) and the crystal structure of bacterial DPP4 bound to the substrate Ile-Pro (PDB ID: 5YP3, orange)26. VbP assumes a pose remarkably like a model substrate. d, NLRP1-CT-DPP9 complex in which DPP9 is coloured by Cα-Cα distances between NLRP1-bound and VbP-bound structures as indicated. A distance scale bar is shown, and VbP is displayed in sticks to mark the active site, with carbon atoms in green, oxygen atoms in red, nitrogen atoms in blue, and boron atoms in orange. NLRP1-CT (UPA) is shown in magenta.
