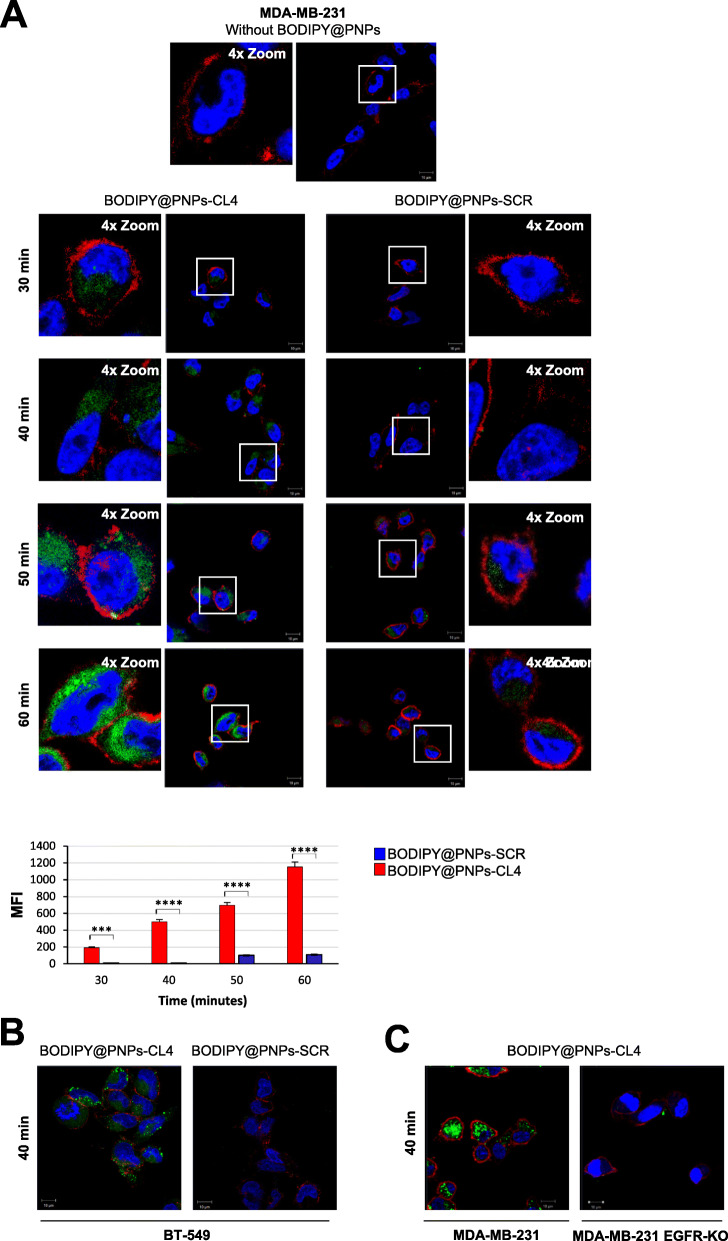
Fig. 2.
Selective cell uptake of BODIPY@PNPs-CL4 compared to BODIPY@PNPs-SCR. (a) Upper, Representative confocal images of MDA-MB-231 cells left untreated or treated with BODIPY@PNPs-CL4 or BODIPY@PNPs-SCR at 37 °C for different periods (from 30 to 60 min). After washing and fixation, cells were labelled with WGA (red) to visualize cell membrane and with DAPI (blue) to stain nuclei. BODIPY@PNPs are displayed in green. White squares indicate the area shown in insets in a magnified view obtained using Image J software. Lower, mean fluorescence intensity (MFI) was evaluated by Zeiss software on a minimum of 50 cells for each sample (n = 3). ****P < 0.0001; ***P < 0.001. (b) Representative confocal images of BT-549 cells treated with BODIPY@PNPs-CL4 or BODIPY@PNPs-SCR at 37 °C for 40 min. (c) Representative confocal images of MDA-MB-231 and MDA-MB-231 EGFR-KO cells treated with BODIPY@PNPs-CL4 at 37 °C for 40 min. (a-c) Magnification 63×, 1.0× digital zoom, scale bar = 10 μm. All digital images were captured at the same setting to allow direct comparison of staining patterns