Figure 2.
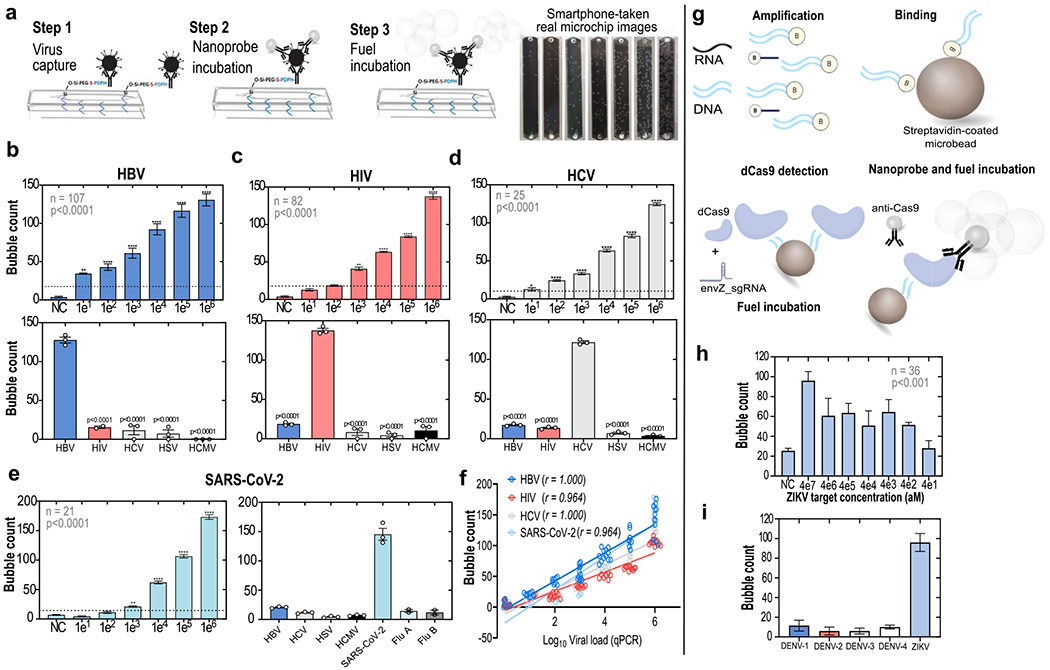
Standardization of the microfluidic chip-based assays to detect intact viruses and viral nucleic acids. (a) General protocol for intact virus detection on-chip through an enzyme-free optical signal output (bubble generation). The bubble outputs observed in HBV microchips incubated with increasing concentrations of spiked viruses are shown. (b–d) Analytical sensitivities (top panels) and specificities (bottom panels) of the intact virus detection assays for HBV, HIV, and HCV, respectively. (b–d) (Top panels) Mean ± SEM of serial 10-fold dilutions of virus spiked samples (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001). Sample numbers and results of analyses of variance are shown within the charts. (b–d) (Bottom panels) Individually spiked with five different viruses (HBV, HIV, HCV, HSV, HCMV), at concentrations of 1 × 106, and tested in the various assay formats and microchips. (e) Analytical sensitivity and specificity of the assay format to detect the SARS-CoV-2. (f) Bubble counts in the microchip channels were highly correlated with viral loads measured by quantitative real-time PCR (qPCR); r = Spearman’s rho. (g) Translation of the system for viral nucleic acid detection on-chip using CRISPR/dCas9. Shown are mean ± SEM. (h) Sensitivity of detection of a ZIKV synthetic genomic region; aM = attomolar. (i) Specificity of the ZIKV nucleic acid detection assay.
