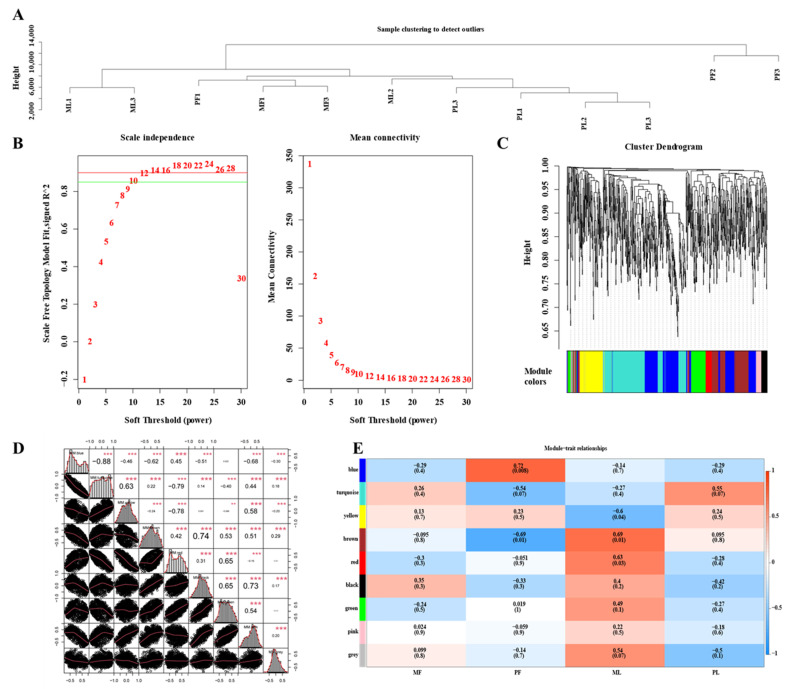
Figure 5.
(A) Dendrogram of all 12 samples; (B) Soft threshold determination of gene co-expression network. The left panel shows the scale-free fit index (y-axis) as a function of the soft-thresholding power (x-axis). The right panel displays the mean connectivity (degree, y-axis) as a function of the soft-thresholding power (x-axis). (C) Shows a hierarchical cluster tree of coexpression modules identified by WGCNA. Every leaf on a tree is a gene. The main branches are made up of 9 modules, which are color-coded. (D) ME correlation between different modules. Black dots represent circRNAs, and the higher the value, the higher the correlation. The more and larger the asterisk, the greater the pairwise correlation between the modules. (E) Association analysis of gene co-expression network modules with tissues. Each row corresponds to a module, whose name is displayed on the left, and each column corresponds to a particular sample. The color of each cell at the row-column intersection indicates the correlation coefficient between the module and the sample. High correlations between specific modules and samples are shown in red.