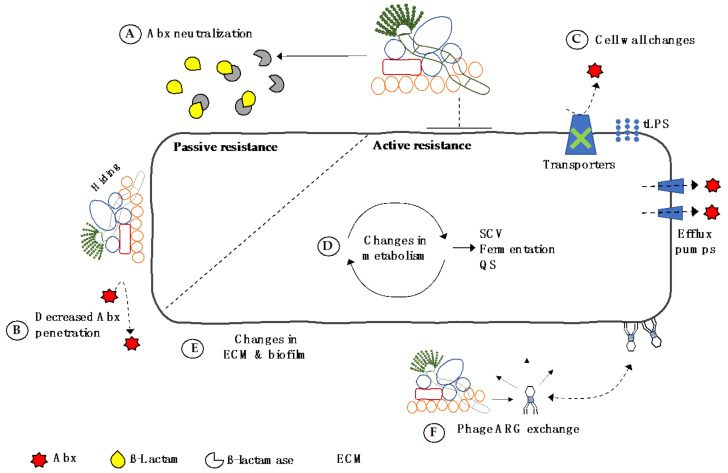
Figure 2.
Passive and active mechanisms of antibiotic tolerance/resistance developed by P. aeruginosa during polymicrobial interactions. (A) Neutralisation of antibiotics by cleaving enzymes produced by other community members; (B) ‘hiding’ in multispecies biofilms reducing access of antibiotics; (C) changes in cell wall architecture, including truncated LPS, increased expression of efflux pumps, or reduced activity of membrane transporters; (D) changes in metabolism resulting in alterations to growth and quorum sensing, development of SCVs, and conversion to fermentative growth; (E) increased or decreased production of ECM and biofilm growth; and (F) transfer of mobile ARGs via bacteriophage. Abx = antibiotics, ECM = extracellular matrix, SCV = small colony variant, QS = quorum sensing, tLPS = truncated lipopolysaccharide, ARGs = antibiotic resistant genes.