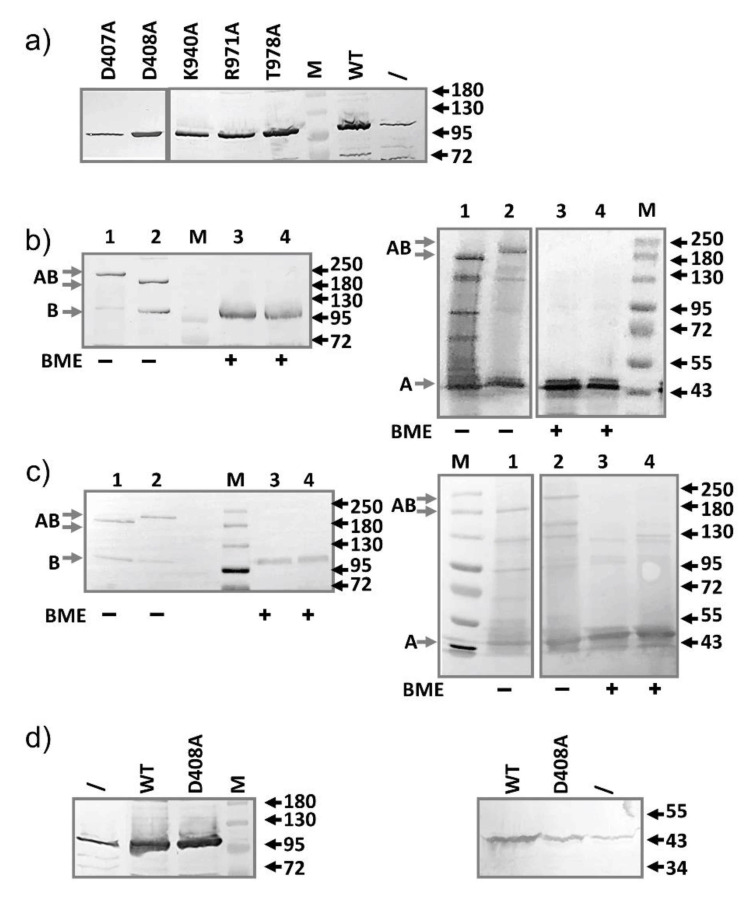
Figure 1.
Characterization of mutants defective in the proton translocation pathway. (a) The point mutation did not affect expression level. Anti-AcrB Western blot analysis of the expression of all five mutants and the wild-type AcrB from plasmid transformed into BW25113. Sample prepared from plasmid-free BW25113 (\) was also prepared and loaded to serve as a control to highlight the difference in expression levels. (b) Anti-AcrB and Anti-AcrA Western blot analyses revealing the formation of disulfide bonded AcrA-AcrB complexes, which was reduced after incubation with BME. AcrA-P57C/AcrB-N191C (lane 1 and 3), AcrA-T217C/AcrB-S258C (lane 2 and 4). (c) Similar to b, with the additional D408A mutation introduced into the constructs. AcrA-P57C/AcrB-N191C/AcrB-D408A (lane 1 and 3), AcrA-T217C/AcrB-S258C/AcrB-D408A (lane 2 and 4). Molecular weight markers are labeled as “M” and the molecular weight of bands (kD) were indicated on the right. The expected bands for AcrA, AcrB, and disulfide bond linked AcrA-AcrB are marked on the left of the gels as A, B, and AB, respectively. (d) Anti-AcrB (left) and anti-AcrA (right) Western blot analysis of BW25113 expressing plasmid pBAD33-AcrAB (WT) or pBAD33-AcrAB-D408A (D408A). Samples prepared from BW25113 not containing plasmid was used as the control (/). For anti-AcrA Western blot, plasmid-containing samples were diluted 4-fold before being loaded into the gel.