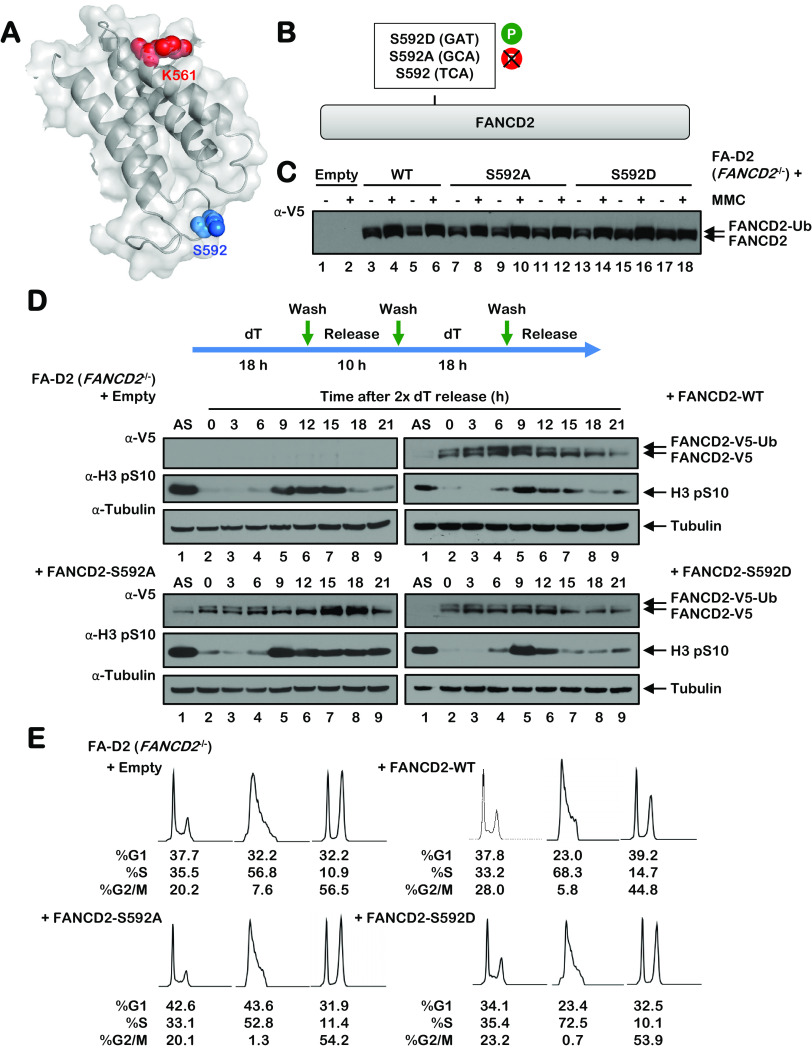
FIG 4.
Mutation of S592 disrupts FANCD2 monoubiquitination during the S phase and following exposure to DNA-damaging agents. (A) Shown is a partial model of the human FANCD2 protein, modeled on the mouse Fancd2 structure (PDB ID 3S4W), illustrating the site of monoubiquitination K561 in red and S592 in blue. (B) Site-directed mutagenesis was used to generate phospho-dead (S592A) and phospho-mimetic (S592D) FANCD2 S592 variants. (C) Polyclonal populations of FA-D2 (FANCD2−/−) patient cells expressing empty vector or V5-tagged FANCD2-WT, FANCD2-S592A, or FANCD2-S592D were incubated in the absence (−) or presence (+) of 200 nM mitomycin C (MMC) for 24 h, and whole-cell lysates were analyzed by immunoblotting. (D) FA-D2 cells stably expressing empty vector or V5-tagged FANCD2-WT, FANCD2-S592A, or FANCD2-S592D were synchronized at the G1/S boundary by double-thymidine block. Cells were released into thymidine-free medium, and whole-cell lysates were analyzed by immunoblotting at the indicated times postrelease. (E) FA-D2 cells stably expressing empty vector or V5-tagged FANCD2-WT, FANCD2-S592A, or FANCD2-S592D were synchronized at the G1/S boundary by double-thymidine block (2× deoxyribosylthymine [dT]) and then released into thymidine-free medium. At the indicated time points, cells were fixed, stained with propidium iodide, and analyzed by flow cytometry. Cell cycle stage analysis was performed using FlowJo v10.2 software.