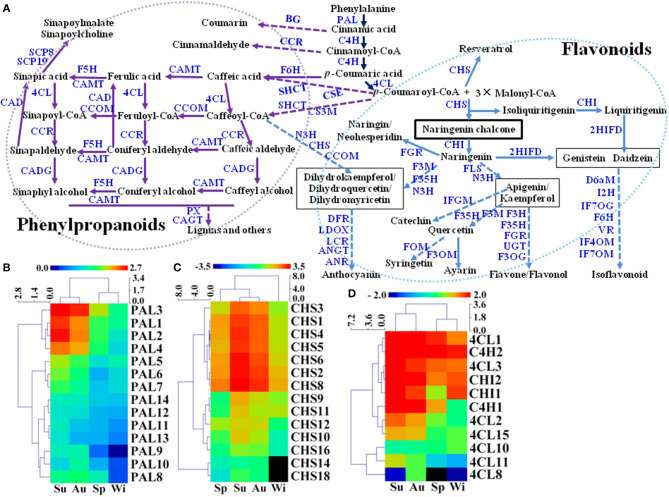
Figure 6.
Putative biosynthetic pathways of flavonoids and phenylpropanoids (FPs) (referring to KEGG pathways map00940 and map00941) in T. hemsleyanum FD-TRs (A) and heatmaps representing expression dynamics of genes involved in phenylalanine ammonia-lyases [PAL, (B)], chalcone synthases [CHS, (C)], 4-coumarate-CoA ligases (4CL), trans-cinnamate 4-monooxygenases (C4H), and chalcone isomerase [CHI, (D)]. 4CL, 4-coumarate-CoA ligase; 2HIFD, 2-hydroxyisoflavanone dehydratase; ANGT, anthocyanidin 5,3-O-glucosyltransferase; ANR, anthocyanidin reductase; BG, beta-glucosidase; C4H, trans-cinnamate 4-monooxygenase; CAD, coniferyl-aldehyde dehydrogenase; CADG, cinnamyl-alcohol dehydrogenase; CAGT, coniferyl-alcohol glucosyltransferase; CAMT, caffeic acid 3-O-methyltransferase; CCOM, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; CHI, chalcone isomerase; CHS, chalcone synthase; CS3M, coumaroylquinate (coumaroylshikimate) 3′-monooxygenase; CSE, caffeoylshikimate esterase; D6aM, 3,9-dihydroxypterocarpan 6a-monooxygenase; DFR, dihydroflavonol-4-reductase; F3M, flavonoid 3′-monooxygenase; F3OG, flavonol 3-O-glucosyltransferase; F3OM, flavonol 3-O-methyltransferase; F35H, flavonoid 3′, 5′-hydroxylase; F5H, ferulate-5-hydroxylase; F6H, flavonoid 6-hydroxylase; FGR, flavanone 7-O-glucoside 2″-O-beta-L-rhamnosyltransferase; FLS, flavonol synthase; FOM, flavonoid O-methyltransferase; I2H, isoflavone 2′-hydroxylase; IF7OG, isoflavone 7-O-glucosyltransferase; IF7OM, isoflavone-7-O-methyltransferase; IF4OM, isoflavone 4-O-methyltransferase; IFGM, isoflavone 7-O-glucoside-6′-O-malonyltransferase; N3H, naringenin 3-dioxygenase; LCR, leucoanthocyanidin reductase; LDOX, leucoanthocyanidin dioxygenase; PAL, phenylalanine ammonia-lyase; PX, peroxidase; SCP8, serine carboxypeptidase-like 8; SCP19, serine carboxypeptidase-like 19; SHCT, shikimate O-hydroxycinnamoyltransferase; UGT, UDP-glucosyl transferase; VR, vestitone reductase.