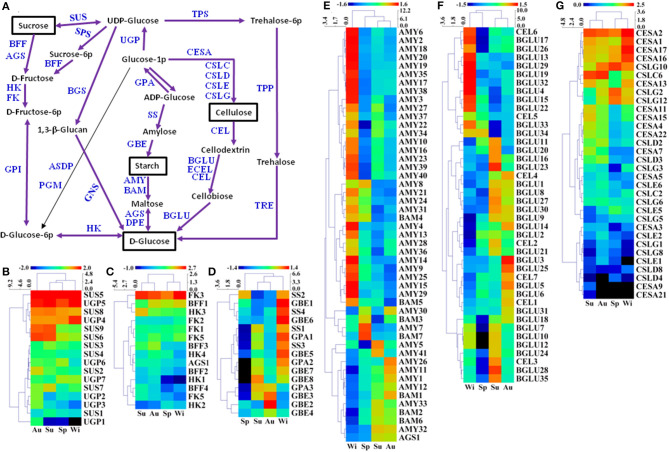
Figure 8.
A putative model of starch and sucrose metabolism (referring to the KEGG pathway map00500) in T. hemsleyanum FD-TRs (A) and heatmaps representing expression dynamics of genes involved in sucrose biosynthesis (B), sucrose degradation (C), starch biosynthesis (D), starch degradation (E), cellulose degradation (F), and cellulose biosynthesis (G). AGS, α-glucosidase; AMY, α-amylase; ASDP, ADP-sugar diphosphatase; BAM, beta-amylase; BFF, beta-fructofuranosidase; BGLU, beta-glucosidase; BGS, 1,3-beta-glucan synthase; CEL, endoglucanase; CESA, cellulose synthase A; CSLC-CSLG, cellulose synthase-like protein C-G; DPE, 4-α-glucanotransferase; ECEL, cellulose 1,4-beta-cellobiosidase; FK, fructokinase; GBE, 1,4-α-glucan branching enzyme; GNS, glucan endo-1,3-beta-glucosidase; GPA, glucose-1-phosphate adenylyltransferase; GPI, glucose-6-phosphate isomerase; HK, hexokinase; PGM, phosphoglucomutase; SPS, sucrose-phosphate synthase; SS, starch synthase; SUS, sucrose synthase; TPP, trehalose 6-phosphate phosphatase; TPS, trehalose 6-phosphate synthase/phosphatase; TRE, α,α-trehalase; UGD, UDP-glucose 6-dehydrogenase; UGP, UTP-glucose-1-phosphate uridylyltransferase.