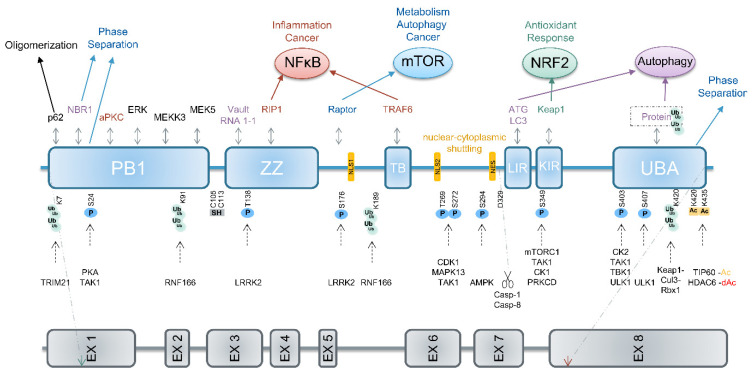
Figure 1.
Structure of SQSTM1 and p62. Domain architecture of p62, posttranslational modifications, binding/modifying proteins and pathways regulated by p62. The PB1 domain mediates oligomerization and activation of p62, UBA-mediated dimerization holds p62 inactive. For phase separation, the PB1 and UBA domains are required. The role of p62 as a cargo receptor in autophagy is mediated by the LIR motif, linking it to LC3 on autophagosomes, and the UBA domain, which binds to ubiquitinated cargos. Upon phosphorylation at Ser349, the KIR motif interacts with Keap1, thereby liberating Nrf2 and inducing Nrf2 target gene expression. When Raptor binds to p62, mTOR is activated inducing metabolic pathways. Binding of RIP1 to the ZZ or TRAF6 to the TB motif supports activation of NF-κB. Cleavage after Asp329 by caspase-8 (casp-8) or caspase-1 (casp-1) generates an amino terminal fragment of p62 activating mTOR. Disulfide bond formation involving Cys105 or Cys113 as well as binding of vault RNA1-1 via the ZZ motif supports autophagy. Please see in the text for details. Abbreviations: Ac: acetylation, dAc: de-acetylation, C: Cys, D: Asp, K: Lys, KIR: Keap1-interacting region, LIR: LC3-interacting region, NES: nuclear export signal/sequence, NLS: nuclear localization signal/sequence, PB1: Phox and Bem1p, S: Ser, T: Thr, TB: TRAF6-binding domain, UBA: ubiquitin-associated, ZZ: Zinc finger, [1,12,13,14].