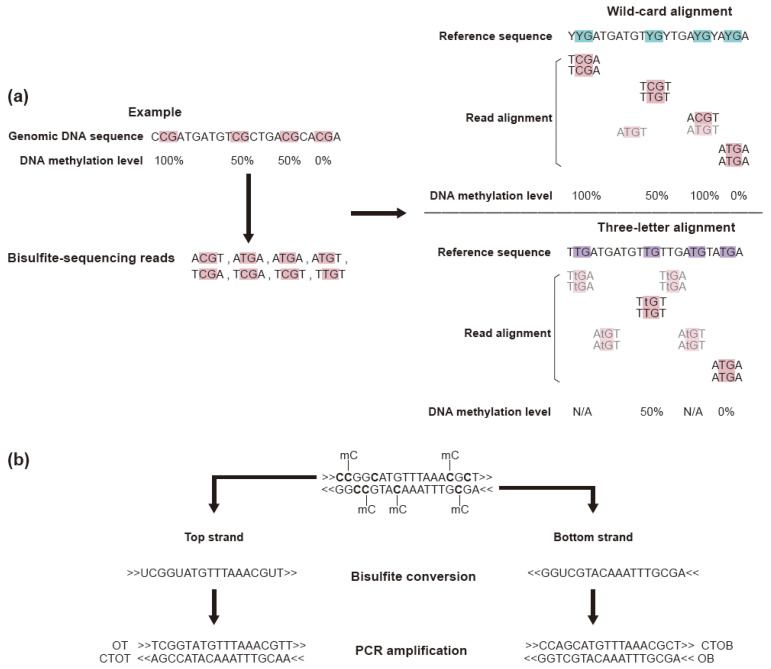
Figure 3.
The bisulfite treatment-based sequencing data analysis should take into consideration the converted normal cytosine. (a) With the exception of the CpG loci (red), all cytosine residues of produced sequence reads are converted to thymine after PCR (left panel). These sequence reads are aligned on the reference genome using the wild-card method (upper right panel) or three-letter method (lower right panel). Although the wild-card alignment aligned more reads and coverage is increased, the methylation level is biased. In the three-letter system, some reads failed to align but the calculated methylation level is unbiased compared to the wild-card method when there is alignment (transparent reads are an alignment failure) (adapted with permission from [82], 2012 Springer Nature). (b) Sequence reads after bisulfite treatment. Due to the PCR step, four types of sequence reads are produced. Two are from the original target molecule (OT, OB), and the other two are from the complementary strand generated by PCR (CTOT, CTOB). OT: original top, OB: original bottom, CTOT: complementary to original top, CTOB: complementary to original bottom (adapted with permission from [83], 2012 Springer Nature).