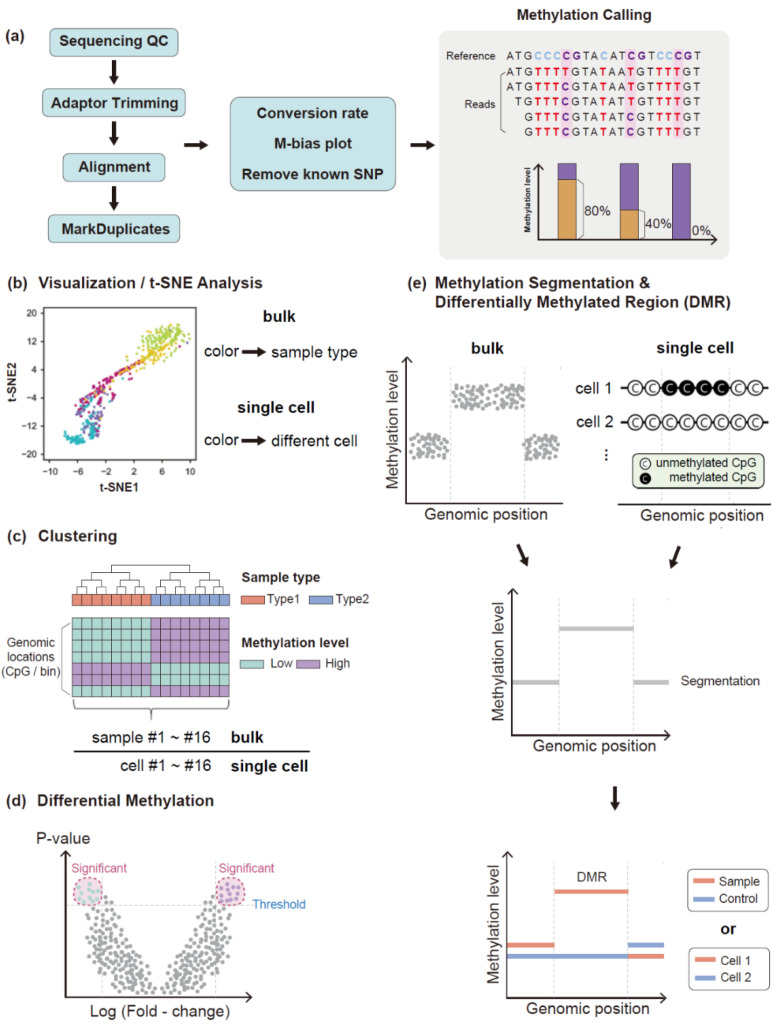
Figure 4.
Analysis methods of DNA methylation. (a) Overview of methylation analysis pipeline. The analysis starts with the quality check of raw sequence reads followed by adaptor trimming and alignment. After alignment, two tracks of analysis are performed. The first is assessment of the experimental quality, such as bisulfite conversion rate, M-bias plot, and removal of known single-nucleotide polymorphisms (SNPs). The second is removal of duplication followed by methylation calling. After methylation calling, several steps, such as visualization (b), cluster analysis (c), and identification of differentially methylated sites or genes between bulk or single-cell groups (d) or regions (e), are carried out. Each analysis method is used on both the bulk scale and single-cell scale, and individual single cells are treated similarly to individual samples in bulk. The image in (b) was adapted from the t-SNE figure of the open access iscCOOL-seq paper (Gu, C., Liu, S., Wu, Q. et al. 2019) [58]. t-SNE: t-stochastic neighbor embedding.