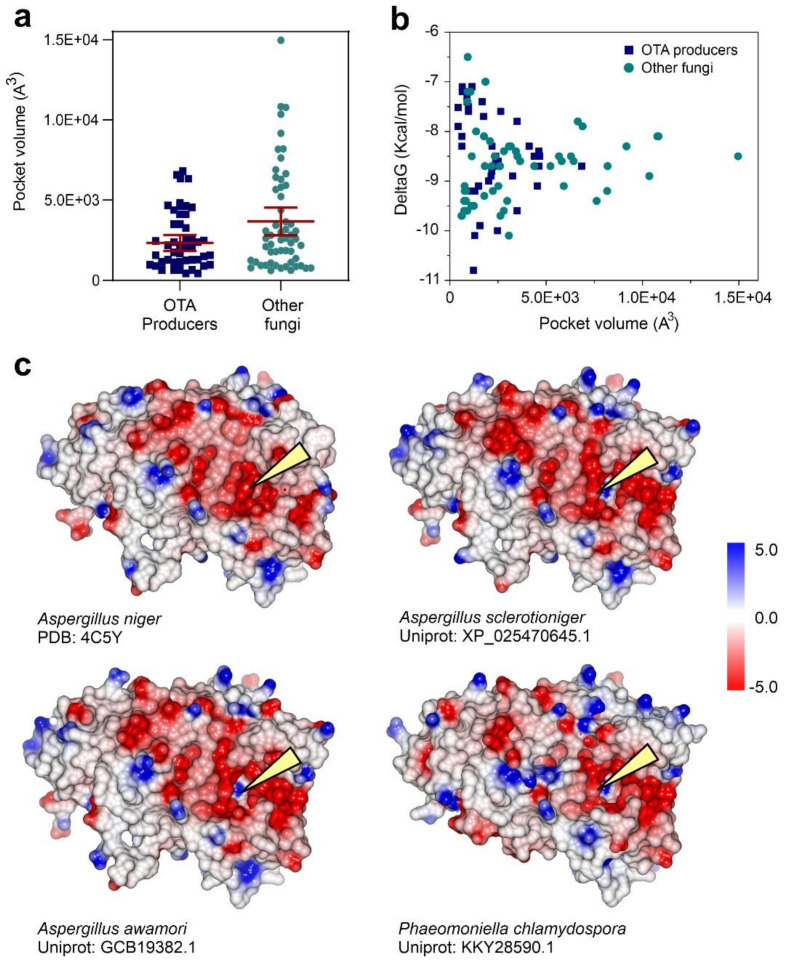
Figure 5.
Characteristics of the substrate-binding pockets in the dataset of fungal ochratoxinase-like enzyme models. (a) Volume of the substrate-binding pocket determined by the CB-Dock algorithm [35], distinguishing between the enzymes encoded by ochratoxin producers and other fungi. (b) Results of the blind docking and pocket search analysis by CB-Dock with OTA as ligand. The plot represents the relationships between the substrate-binding pocket volume in each enzyme model and the calculated Gibbs’ function value for the best docking solution of the ligand. Following the same criteria, the enzyme models are classified according to their original fungal strain. (c) Surface potential representation of selected structural models of the analyzed dataset. The reference structure of A. niger ochratoxinase, together with the enzymes from A. sclerotioniger and A. awamori, are examples of ochratoxinase enzymes from fungal strains characterized as toxin producers, whereas the enzyme from P. chlamydospora is an example of other fungal species not described as ochratoxin producers. The location of the substrate-binding pocket entrance is depicted by a yellow triangle. The electrostatic potential scale is depicted as a gradient color bar.