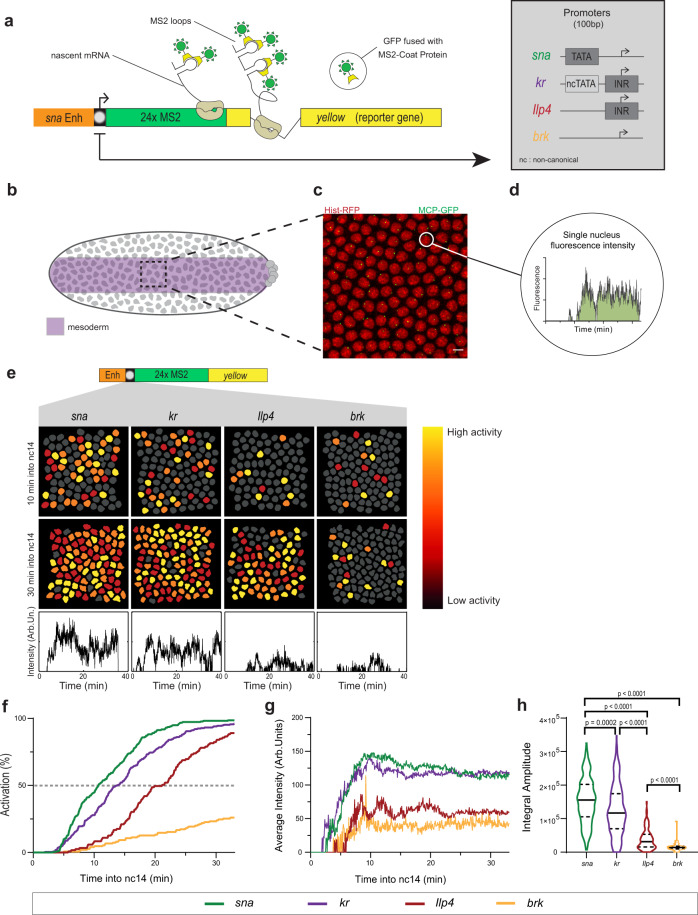
Fig. 1. A synthetic transgenic platform to image promoter dynamics.
a Schematic view of transgenes used to study transcriptional dynamics of sna, kr, Ilp4, and brk core promoters. A minimal sna enhancer was placed upstream of the core promoter followed by 24xMS2 repeats and a yellow reporter gene. Core promoter motifs are indicated in the inset. b Schematic of Drosophila embryo showing spatial restriction of analysis to presumptive mesoderm (purple). c Maximum intensity projection of representative 15 µm Z-stack of snaE < snaPr < 24xMS2-y (snaE < sna) nc14 embryo showing MS2/MCP-GFP-bound transcriptional foci (GFP) and nuclei (His-RFP). Scale bar is 5 µm. d Sample single-nuclei trace showing GFP fluorescence during nc14. The surface of the green region indicates trace integral amplitude. e False-colored frames from live imaging of indicated promoters showing relative instantaneous fluorescence intensity in early and late nc14. Inactive nuclei are gray and highly active nuclei are in yellow. f Cumulative activation curves of all nuclei during the first 30 min of nc14 for sna (green), kr (purple), Ilp4 (red), and brk (blue). Time zero is from anaphase during nc13-nc14 mitosis. g Average instantaneous fluorescence of transcriptional foci of active nuclei during the first 30 min of nc14 for sna (green), kr (purple), Ilp4 (red), and brk (blue). Time zero is from anaphase during nc13-nc14 mitosis. h Distribution of individual trace integral amplitudes from first 30 min of nc14 for sna (green), kr (purple), Ilp4 (red), and brk (blue). The intensity amplitude at a given time may result from the overlap of several bursts and is a convolution of promoter active/inactive times, polymerase initiation frequency, and the duration of a single polymerase signal. The integral amplitude estimates the transcriptional activity and the total number of transcripts at a steady state; it is proportional to the probability of active state (pON) initiation rate (kINI) and to the duration of the signal. Solid lines represent median and dashed lines first and third quartiles, using a one-tailed Kruskal–Wallis test for significance with multiple comparison adjustments. Statistics: snaE < sna, 216 nuclei, 3 movies; snaE < kr, 243 nuclei, 4 movies; snaE < Ilp4, 114 nuclei, 2 movies; snaE < brk, 45 nuclei, 2 movies. See Supplementary Movies 1 and 5.