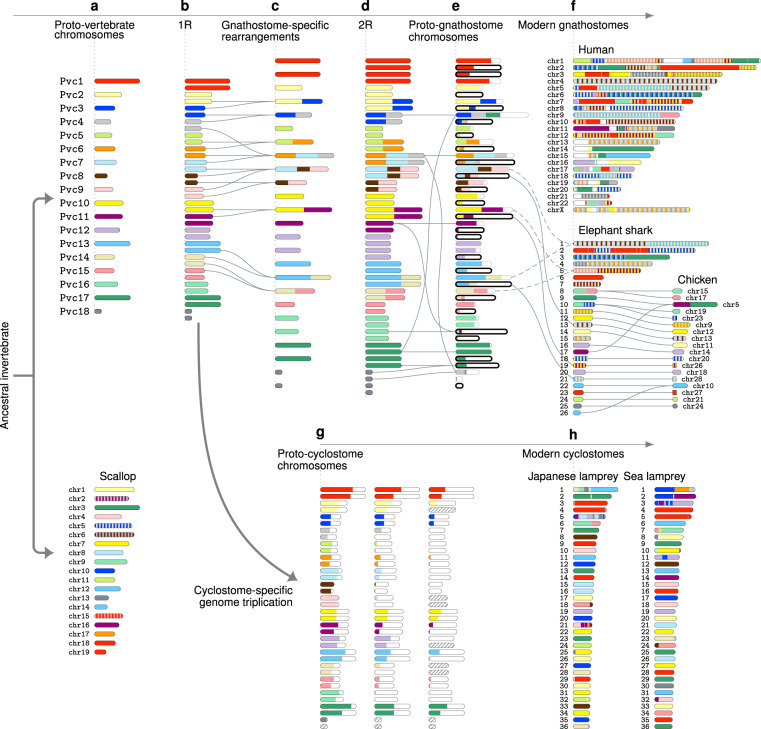
Fig. 6. Inferred scenario of the early vertebrate genome evolution.
a The 18 proto-vertebrate chromosomes show strong macrosynteny conservation in the scallop genome. Scallop chromosomes are coloured to indicate homology with proto-vertebrate chromosomes, where stripes indicate homology to multiple proto-vertebrate chromosomes. b The first tetraploidization (1R) doubled the proto-vertebrate chromosomes. c The post-1R genome experienced nine chromosome fusions, which were not shared with the proto-cyclostome lineage. d The second tetraploidization (2R) established the proto-gnathostome genome with 49 chromosomes after a few post-2R fusions. e The duplicated proto-gnathostome chromosomes exhibit biased gene loss/retention, as indicated by different lengths of coloured bars (see Supplementary Note 4 for details). Chromosomes surrounded by a thick line belong to the shorter subgenome (with a higher rate of gene loss). f Chromosomal regions in modern gnathostome genomes were painted according to the originating proto-gnathostome chromosomes, where stripes indicate fusion chromosomes in the proto-gnathostome genome (e.g. human chr9 was derived from Pvc13, 14 and 15). The largest 26 elephant shark scaffolds and selected chicken chromosomes demonstrate the persistent conservation of microchromosomes. The 2R event duplicated fused chromosomes, each of which gave rise to a large chromosome (dashed lines) and a microchromosome (solid lines) in elephant shark and chicken, which suggests that microchromosomes were created by biased gene fractionation after 2R. g The cyclostome lineage diverged from the gnathostome lineage shortly after 1R, and the proto-cyclostome genome was formed by hexaploidization. Coloured bars show biased gene loss/retention, and chromosomes missing in our reconstruction are shown as hatched bars. h The largest 36 scaffolds in modern lamprey genomes were painted according to their originating proto-vertebrate chromosomes.