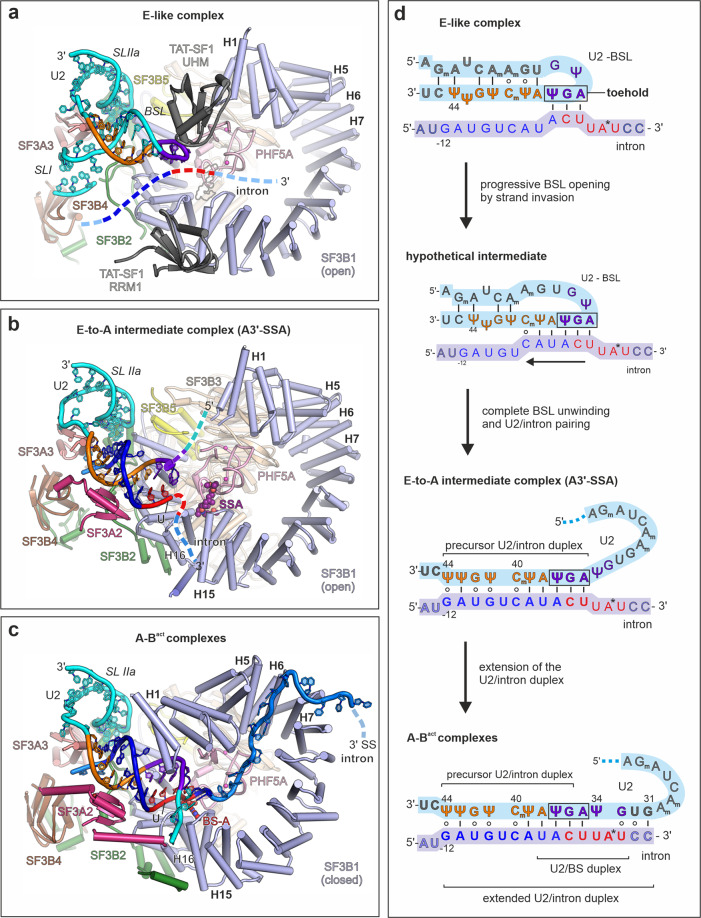
Fig. 6. Mechanism of the stepwise intron selection by U2 snRNP.
a The E-like complex model is based on the 17S U2 snRNP structure (PDB 6Y50), which is positioned in the proximity of an intron schematic. The equivalent location of SSA is shown as a silhouette, for orientation’s sake. b Structure of the U2 5′ module of the A3′ complex arrested with spliceostatin A (SSA). c Structure of the U2 5′ module from the A/Bact complexes. The depicted U2 5′ model is derived from the human Bact coordinates (PDB 5Z58). Complexes from a to c were superimposed over equivalent residues from PHF5A and SF3B1. d Schematic RNA transitions during intron’s recognition by U2, as suggested by cryo-EM structures. Nucleotides resolved in cryo-EM structures are shown in bold letters and colored as in a. Ψ and the “m” subscript denotes pseudouridine and methylated nucleobases, respectively. Throughout the entire figure, the BS consensus and nucleotides from BSL’s loop are shown in red and purple, respectively. The remaining sequence of the intron and of the U2 snRNA are colored in deep blue and orange, respectively. RNA strands not resolved in cryo-EM structures are dashed. All the other subunits and elements are color-coded as in Fig. 1.