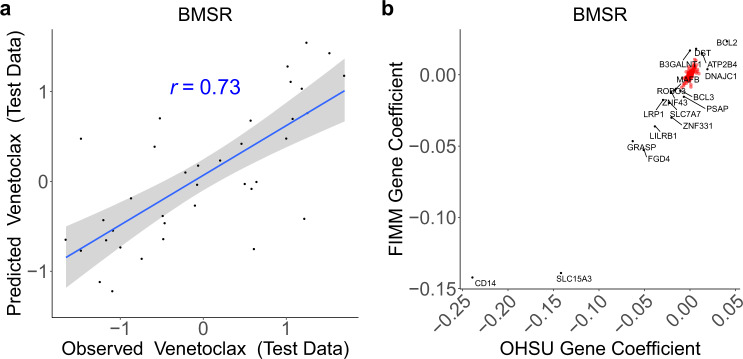
Fig. 5. Integrative analysis reveals monocyte-associated biomarkers predictive of venetoclax resistance.
a Observed (x axis) versus BMSR-predicted (y axis) venetoclax response. Expression-based Bayesian regression model trained using fivefold cross-validation on combined OHSU and FIMM datasets (n = 159) and tested on held-out fold yielding median performance across the fivefolds (n = 37). b Coefficients of genes (n = 2132) in OHSU (x axis) or FIMM (y axis) datasets following training of Bayesian regression modeling simultaneously on both datasets (n = 196). r: Pearson correlation; dashed line: identity line; blue line: linear regression fit; gray shading: 95% confidence interval. Labeled genes were those having extremal (Stouffer’s p < 0.01) combined coefficients across both datasets.