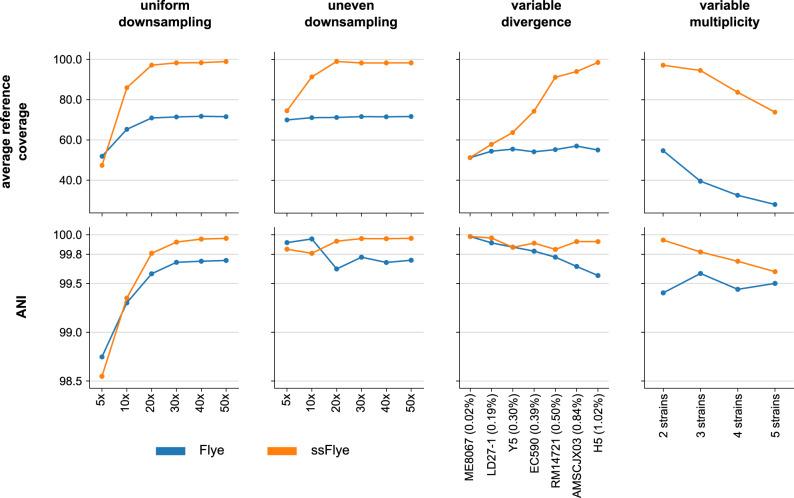
Fig. 4. Evaluation of strain-separated assemblies with respect to strain coverage, divergence, and number of strains.
Average reference coverages and nucleotide identities of the strain-oblivious Flye assemblies and the Strainberry separated assemblies (ssFlye) on simple mock communities characterized by variable strain coverage (first two columns), divergence (third column), and number of strains (fourth column). The variable coverage communities are downsampled versions of the Mock3 dataset (B. cereus, E. coli strain K-12, and E. coli strain W). We kept the same depth of coverage among the strains (uniform downsampling) or a constant 50× coverage for B. cereus and E. coli strain K-12 while downsampling exclusively E. coli strain W (uneven downsampling). The variable divergence datasets consist of simulated reads from two strains where one is E. coli strain K-12 and the other one is listed on the x-axis (with the divergence percentage shown in parenthesis). The datasets with variable number of strains contain 2, 3, 4, and 5 conspecific strains of E. coli with pairwise divergences ranging from 0.7% to 1.4%.