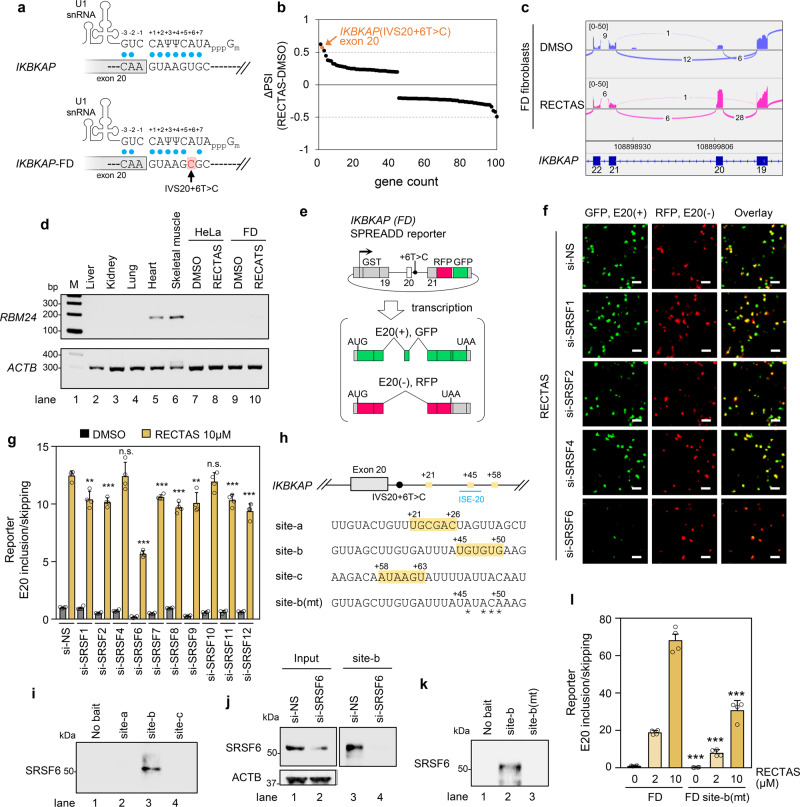
Fig. 1. IKBKAP-FD exon 20 requires SRSF6 binding to an ISE.
a The IKBKAP exon 20 5′ splice site has nearly optimal pairing to U1 snRNA, whereas the IVS20 + 6 T > C creates an additional mismatch at +6 position, resulting in suboptimal recognition by U1 snRNP. Blue dots indicate base pairing. b Top 100 differential splice events by | ΔPSI (PSIRECTAS-PSIDMSO) | following RECTAS treatment (2 µM, 8 h) in FD patient fibroblasts. c Sashimi plot with junction read number of IKBKAP-FD exon 19–22 for DMSO- or RECTAS-treated FD patient fibroblasts. The displayed read count range was set to 0–50. d RT-PCR for RBM24 expression in human tissues, and cell lines treated with 10 µM RECTAS or 0.1% DMSO for 24 h. RBM24 and ACTB were detected with primers oAM669 + oAM671 and oAM13 + oAM14, respectively. e Diagram of reporter system for IKBKAP-FD exon 20 splicing13,17. GFP is translated when exon 20 is included (E20(+)), whereas RFP is translated when it is skipped (E20(−)). f, g Microscopic images (f) and quantification of IKBKAP-FD exon 20 inclusion (g), following treatment with RECTAS (10 µM) or 0.1% DMSO for 24 h in HeLa cells transfected with the indicated siRNAs for 72 h. h SRSF6-binding motifs starting at +21 nt (site-a), +45 nt (site-b), and +58 nt (site-c), predicted by ESE finder 3.028,29 are shown. RNA sequences for pull-down assays are indicated. Asterisks in site-b(mt) indicate mutated ribonucleotides. ISE-20 from Sinha et al.30 is also indicated. i–k Western blot of SRSF6 by 1H4 antibody for RNA pull-down products or no bait RNA control for HeLa cells (i), site-b pull-down products for HeLa cells treated with siRNA for 72 h (j), and site-b and site-b(mt) RNA pull-down products for HeLa cells (k). l Quantification of IKBKAP exon 20 inclusion rate, following RECTAS treatment for 24 h in HeLa cells with IKBKAP-FD or site-b(mt)-mutant IKBKAP (FD site-b(mt)) reporter. Data from four replicates are shown in (f), (g), and (l), and two replicates are shown in (i), (j), and (k). Representative data of two experiments are shown in (d). Columns, mean; bars, SE; n.s., P ≥ 0.05; **P < 0.01; ***P < 0.001 for unpaired two-tailed t test in (g) (vs. the RECTAS 10 µM of si-NS: si-SRSF1, P = 2.3 × 10−3; si-SRSF2, P = 1.7 × 10−4; si-SRSF4, P = 0.94; si-SRSF6, 1.5 × 10−7; si-SRSF7, P = 1.9 × 10−4; si-SRSF8, P = 1.0 × 10−4; si-SRSF9, 3.0 × 10−3; si-SRSF10, P = 0.28; si-SRSF11, P = 6.1 × 10−4; si-SRSF12, P = 1.8 × 10−4) and (l) (vs. the FD of corresponding RECTAS concentration: 0 µM, P = 2.0 × 10−4; 5 µM, P = 4.4 × 10−5; 10 µM, P = 1.0 × 10−4).