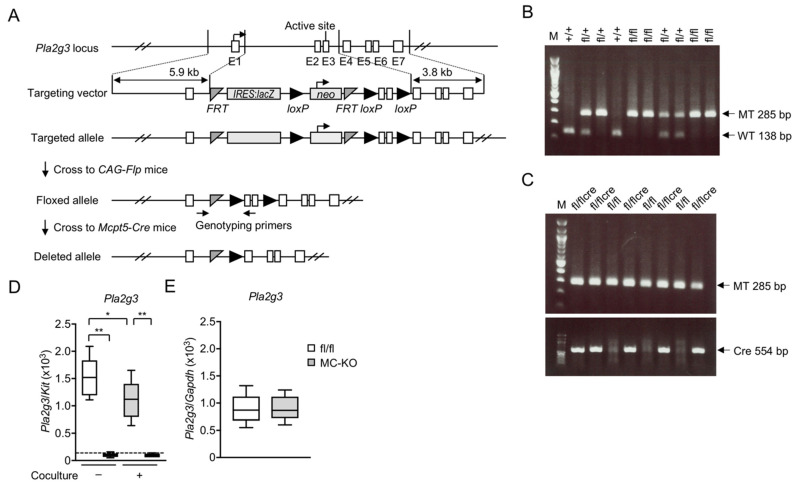
Figure 1.
Generation of MC-specific Pla2g3-deficient mice. (A) A schematic representation of the mouse Pla2g3 locus, the targeting vector from KOMP, the targeted Pla2g3 allele with a lacZ/neo cassette, the floxed Pla2g3 allele after removal of the lacZ cassette by flippase, and the Pla2g3-deleted allele after removal of the neo cassette by Cre recombinase. The floxed mice were bred with Mcpt5-Cre mice, and sequences between the two loxP sites were removed from the offspring’s genome in an MC-specific manner. Positions of primers for genotyping are marked with arrows. E1–7 (open boxes), Pla2g3 exons; FRT (gray isosceles triangles), sites for flippase; IRES, internal ribosome entry site; lacZ (gray box), gene encoding β-galactosidase; neo (gray box), neomycin phosphotransferase; loxP sites (black right triangles), target sites for Cre recombinase. Adapted from www.KOMP.org. (B,C) A representative genotyping PCR on agarose gels. The amplified PCR products specific for the floxed allele (MT, 285 bp), WT Pla2g3 allele (WT, 138 bp), and Mcpt5-Cre allele (Cre, 554 bp) are indicated. (D,E) Quantitative RT-PCR of Pla2g3 in BMMCs with (+) or without (−) coculture with 3T3 fibroblasts (n = 6) (D) and splenocytes (n = 8) (E) from Pla2g3fl/fl (fl/fl) and Pla2g3fl/flMcpt-5-Cre mice (MC-KO). The dotted line is the threshold for the detection limit. Data are presented as box plots with Tukey whiskers. *, p < 0.05; **, p < 0.01; one-way ANOVA (D); Mann–Whitney test (E). Data are pooled from 2 independent experiments, each of which gave similar results.