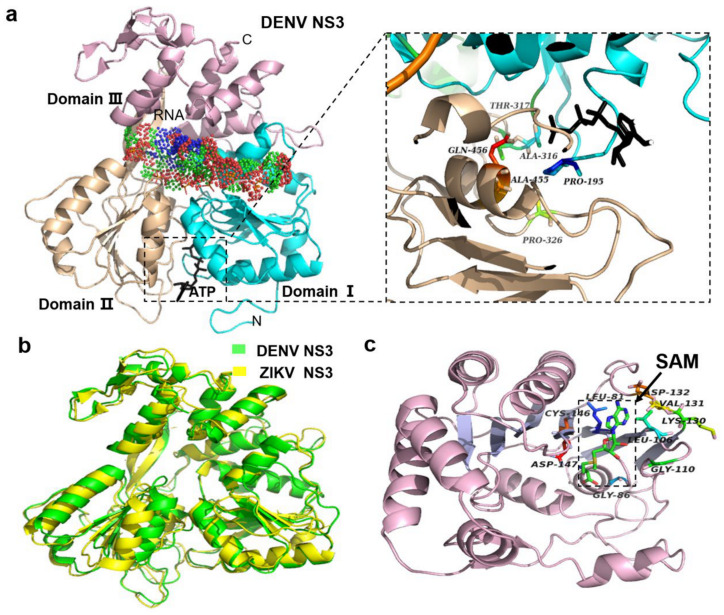
Figure 4.
The structures of non-structural protein 3 (NS3) and NS5 of flaviviruses. (a) NS3 protein structure of dengue virus (DENV; Protein Data Bank [PDB]: 2JLV). The gap between the three domains (indicated by three different colors) is the RNA-binding site, and the adenosine triphosphate (ATP)-binding site is between domains I and II (see the enlargement of the framed area). (b) NS3 of Zika virus (ZIKV) and DENV. The root mean square (RMS) of the two viruses is 1.43, indicating that they are very similar. According to this reason, some proteins of other flaviviruses are also similar, which is very helpful for structural development and vaccine design (PDB: 2JLR DENV; PDB: 5JRZ ZIKV). (c) In the methyltransferase (MTase) core of NS5, there is a binding site of S-adenosylmethionine (SAM) (green in the framed area, indicated by an arrow). This site is also termed AdoMet, and plays an important role in the RNA capping process. This site is wrapped in the hydrophobic pocket of the central split (PDB: 2WA2 NKV).