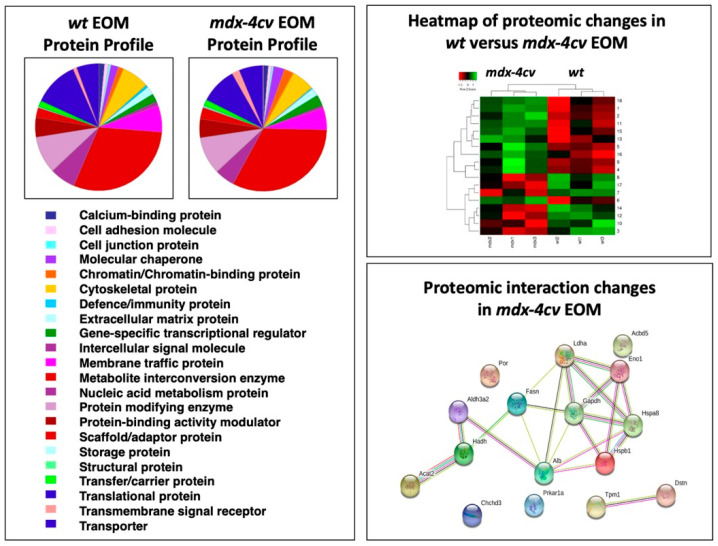
Figure 4.
Bioinformatic analysis of the comparative proteomic profiling of extraocular muscle (EOM) from wild type (wt) versus the mdx-4cv model of X-linked muscular dystrophy. Shown is the result of the bioinformatic PANTHER analysis [46] of the distribution of protein classes within the EOM proteome from normal versus dystrophic mice. In addition, the heat map of the comparative proteomic analysis of wt versus mdx-4cv EOM is displayed, which shows the findings from hierarchical clustering of the mean protein expression values of statistically significant differentially abundant EOM proteins. Potential changes in protein–protein interaction patterns in mdx-4cv EOMs were determined with the help of the bioinformatics software programme STRING [47].