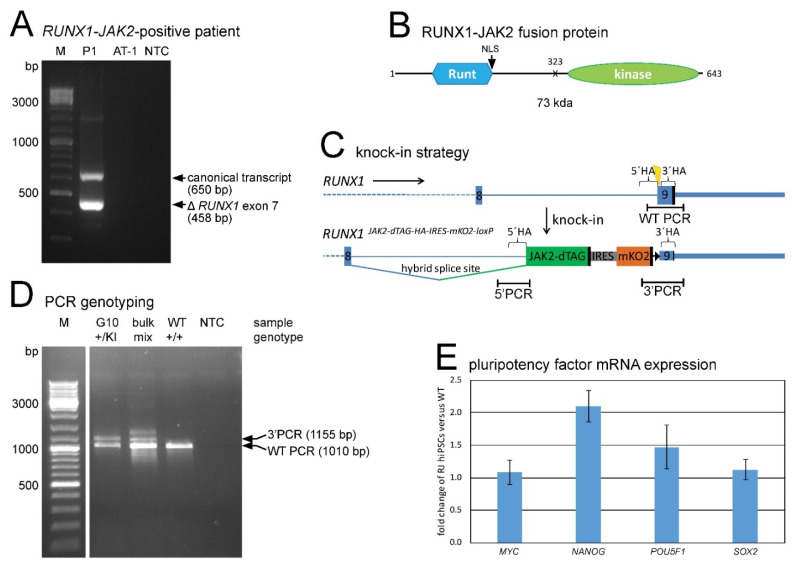
Figure 1.
RUNX1-JAK2 fusion gene identification and establishment of corresponding knock-in hiPSC lines. (A) Reverse transcription polymerase chain reaction (RT-PCR) with RUNX1 forward and JAK2 reverse primers showed expression of two in-frame fusion transcripts (the canonical full-length and a splice variant lacking RUNX1 exon 7) in patient P1. The ETV6-RUNX1-positive cell line AT-1 and a no template control (NTC) served as negative controls (bp, base pair). (B) Putative RUNX1-JAK2 protein structure depicted with the Runt homology domain and the nuclear localization signal (NLS) of RUNX1 and the JH1 tyrosine kinase domain of JAK2, the breakpoint (X), the amino acid positions, and the expected molecular weight (kda, kilodalton). (C) Knock-in strategy: The JAK2 encoded part fused to a dTAG, and an IRES-mKO2 cassette were inserted into RUNX1 exon 9. Locations of CRISPR/Cas9 target site (yellow bolt), homology arms (5′HA and 3′HA), PCR amplicons (5′ and 3′ PCR detecting the knock-in flanks, and WT PCR the wild-type allele) and stop codons (black vertical bars) are shown. A loxP site (black triangle) persisted after Cre-mediated excision of the puromycin resistance cassette (not shown). (D) Three primer PCR of a knock-in clone example (G10) and a cell bulk yielded WT and 3′ PCR products (WT, parental cell line; NTC, no template control). Deduced genotypes are indicated (+, wild-type allele; KI, knock-in allele; mix, +/KI mixed with +/+ or +/floxed). (E) Expression of the four indicated pluripotency factors in RUNX1-JAK2 hiPSCs was determined by quantitative RT-PCR and normalized to GUSB and ABL1. Fold expression of 8 different knock-in lines (RJ, n = 8; mean ± standard deviation) compared to wild-type (WT) hiPSCs is shown.