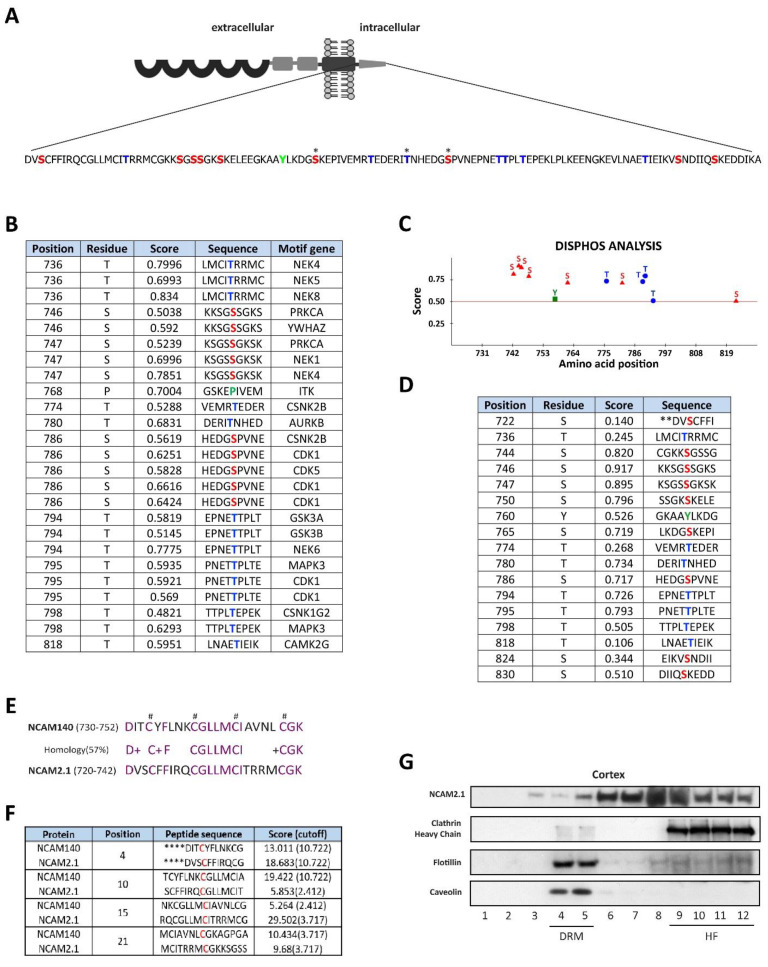
Figure 1.
In silico analysis of NCAM2.1 cytoplasmatic domain. (A) Schematic representation of phosphorylation sites in NCAM2.1 cytoplasmatic tail amino acid sequences. Serine (red), Tyrosine (green) and Threonine (blue) phosposites are represented. * phosphorylated sites previously described. (B) In silico analysis using Scansite 4.0 to identify motifs gene interactors (motif gene) that are likely to be phosphorylated by specific protein kinases or binding domains such as SH2 domains, 14-3-3 domains or PDZ. (C,D) Schematic representation and Table of phosphorylation sites of NCAM2 cytoplasmatic tail with their in silico predicted scores using Disphos. (E) Comparison of NCAM140 (730–752 amino acids) and NCAM2.1 (719–741 amino acids) sequences, NCAM1 has an intracellular region with palmitoylation modification sites that contain cysteine-residues which are critical for its localization in lipid rafts and also present in NCAM2.1 cytoplasmatic domain (#). (F) In silico analysis of NCAM2.1 and NCAM1 with their predicted scores using CSS-Palm software 4.0. NCAM2.1 contains the same potential palmitoylation sites similarly to NCAM1 with high scores (G) WB of NCAM2.1 from cortical extracts subjected to sucrose gradient. NCAM2.1 co-signals in lipid rafts (lanes 4–5, DRM, Detergent-resistant membrane, identified by Flotillin and Caveolin), but mainly outside lipids rafts (lanes 6–12, HF, High Fraction).