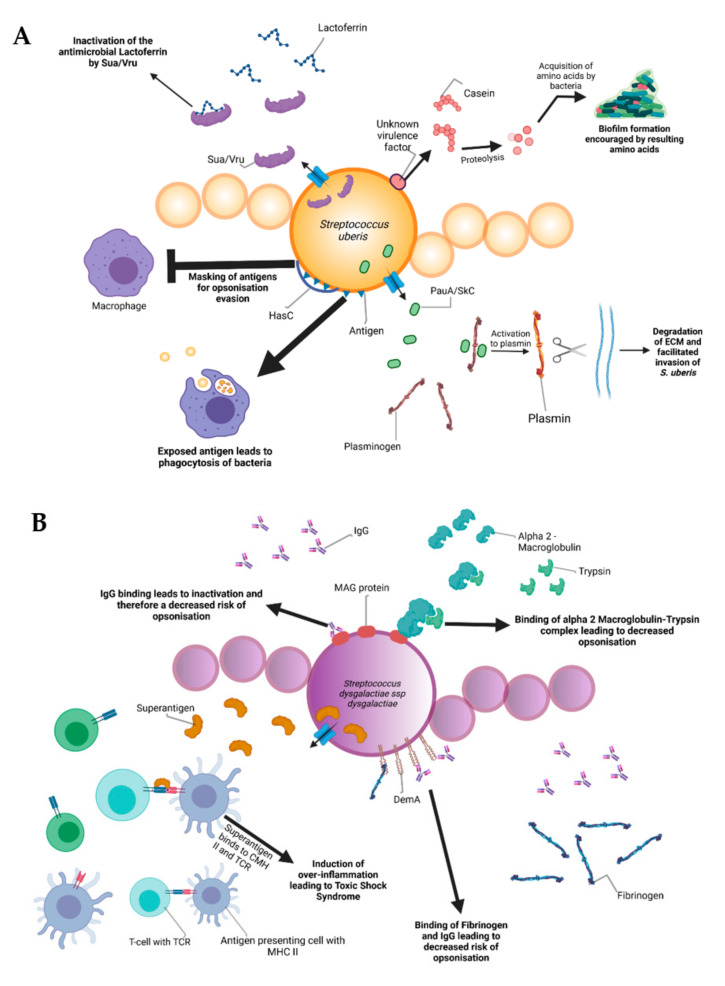
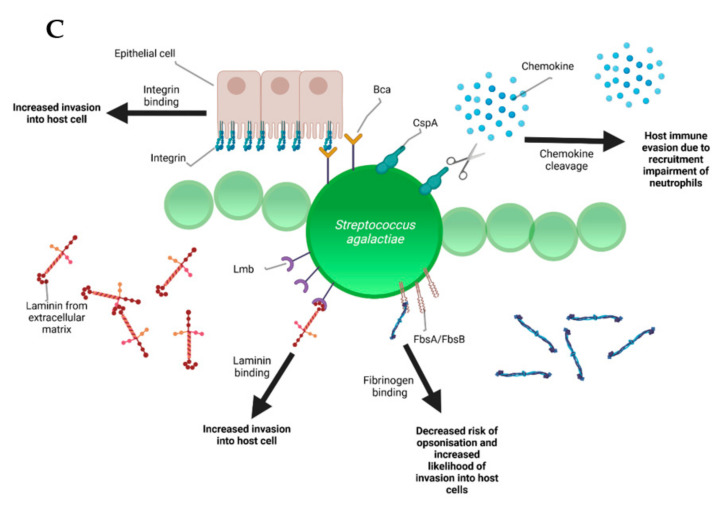
Figure 2.
Schematic representation of the bovine-associated streptococcal virulence factors. (A) S. uberis harbors the capacity for preventing opsonization via the synthesis of the protein HasC, capable of masking the antigens present on the surface of S. uberis. Synthesis of Sua and Vru proteins allow binding of the antimicrobial lactoferrin and thus creating a less hostile environment for the pathogen. Lysis of casein through a so far unknown process provides extra nutrients and thereby favors biofilm formation. PauA and SkC proteins confer plasminogen-activating capabilities to S. uberis. The activation of plasminogen to plasmin leads to the degradation of the extracellular matrix, which in turn facilitates bovine mammary epithelial cell invasion. (B) S. dysgalactiae is equally capable of avoiding opsonization via the binding of fibrinogen with the M-like protein DemA, the binding of IgG and the binding of the alpha 2 macroglobulin-trypsin complex via the synthesis of the MAG protein. (C) S. agalactiae possesses virulence factors implicated in invasion of host cells, namely Bca for integrin binding and Lmb for laminin binding. Immune evasion is facilitated via the synthesis of proteins such as FbsA and FbsB implicated in fibrinogen binding. This leads to a decreased risk of opsonization by phagocytic cells. Synthesis of the serine protease CspA allows S. agalactiae to cleave certain chemokines, which are responsible for neutrophil recruitment, further increasing immune evasion potential.