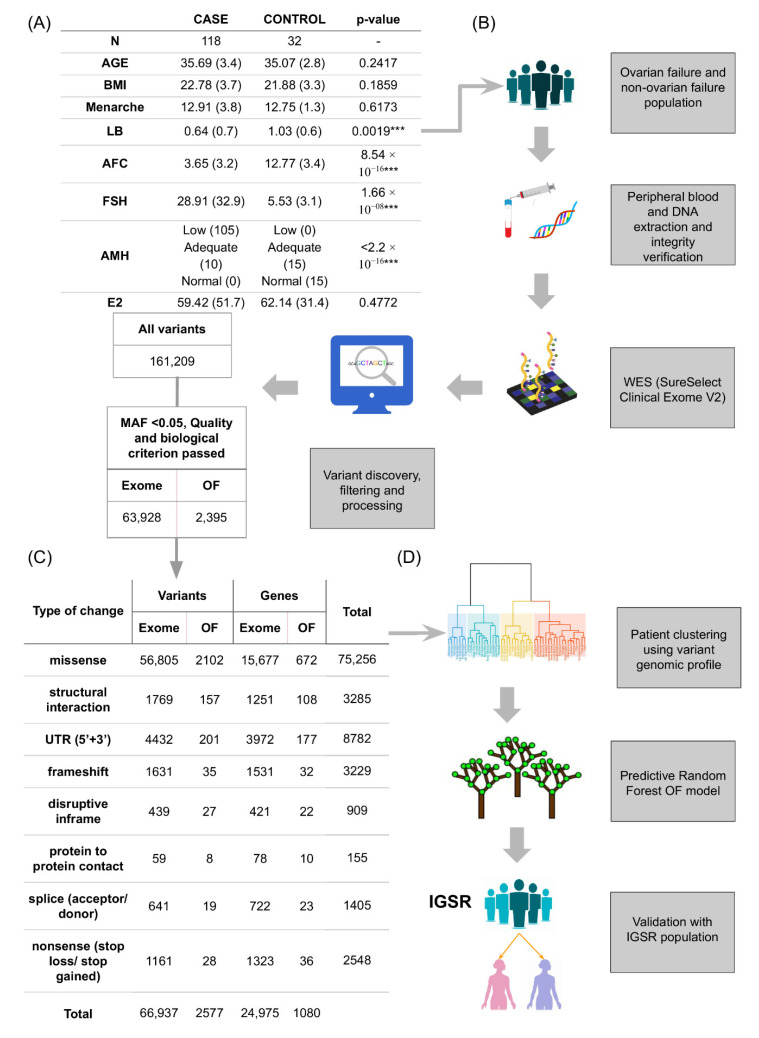
Figure 1.
Study design and variant prioritisation. (A) Population demographics and clinical data. Means and standard deviations (in brackets) are shown. For age, body mass index (BMI), age at menarche, live birth (LB), antral follicle count (AFC), follicle stimulating hormone (FSH), and oestradiol (E2) contrasts. The Shapiro–Wilk test was used to check normality and the Wilcoxon test to evaluate differences between cases and controls. The Fisher test was used to evaluate differences between cases and controls for anti-Müllerian hormone (AMH) levels according to Reference Laboratory hormonal ranges (low: <0.68 ng/mL; adequate: 0.68–2.27 ng/mL; normal: >2.27 ng/mL) (*** p < 0.01). (B) Pipeline for filtering variants. Women diagnosed with ovarian failure (OF) were recruited as cases (n = 118) and those without ovarian failure as controls (n = 32). Whole-exome sequencing (WES) of DNA from peripheral blood was performed in all samples using SureSelect Clinical Research Exome V2 (Agilent Technologies) and Illumina sequencing (Miseq or Nextseq). Variant calling was performed using GATK software. (C) Prioritised variants. Variants that passed quality and biological criteria (minor allele frequency (MAF), type of change at protein level, sequencing parameters) are shown for targeted analysis of genes previously associated with ovarian physiology and from whole exome analysis. Number of variants and genes affected for each predicted change in protein function are also represented. (D) Pipeline of the predictive OF model. A random forest predictive model is built using the prioritised variants and then validated with the pseudo-control population of the International Genome Sample Resource (IGSR) (n = 1271).