Abstract
Solitary fibrous tumor is a rare subtype of soft-tissue sarcoma with a wide spectrum of histopathological features and clinical behaviors, ranging from mildly to highly aggressive tumors. The defining genetic driver alteration is the gene fusion NAB2–STAT6, resulting from a paracentric inversion within chromosome 12q, and involving several different exons in each gene. STAT6 (signal transducer and activator of transcription 6) nuclear immunostaining and/or the identification of NAB2–STAT6 gene fusion is required for the diagnostic confirmation of solitary fibrous tumor. In the present study, a new gene fusion consisting of Nuclear Factor I X (NFIX), mapping to 19p13.2 and STAT6, mapping to 12q13.3 was identified by targeted RNA-Seq in a 74-year-old female patient diagnosed with a deep-seated solitary fibrous tumor in the pelvis. Histopathologically, the neoplasm did not display nuclear pleomorphism or tumor necrosis and had a low proliferative index. A total of 378 unique reads spanning the NFIXexon8–STAT6exon2 breakpoint with 55 different start sites were detected in the bioinformatic analysis, which represented 59.5% of the reads intersecting the genomic location on either side of the breakpoint. Targeted RNA-Seq results were validated by RT-PCR/ Sanger sequencing. The identification of a new gene fusion partner for STAT6 in solitary fibrous tumor opens intriguing new hypotheses to refine the role of STAT6 in the sarcomatogenesis of this entity.
Keywords: solitary fibrous tumor, gene fusion, STAT6, NFIX–STAT6
1. Introduction
Solitary fibrous tumor (SFT) is a rare and ubiquitous subtype of soft-tissue sarcoma (STS) that was originally described in pleural tumors by Klemperer and Rabin in 1931 [1,2]. These fibroblastic tumors of mesenchymal origin have an incidence of one new case per million people each year, exhibiting distinct clinical behaviors, from mildly to highly aggressive neoplasms. In line with this, the last WHO classification recommended instead of “typical SFT” or “malignant SFT” terms, the use of risk-stratification models as a better tool to determine prognosis in these tumors [3,4]. The percentage of recurrence into advanced disease can be as high as 40%, and even higher in series with a long follow-up period [5,6,7].
The histopathology of SFT displays a wide range of features, such as a pattern of irregularly arranged spindle to ovoid cells along with other components, staghorn-shaped blood vessels and a prominent collagenous stroma. The pathological spectrum can vary from highly fibrotic with scanty cells to highly cellular, as well as the dedifferentiated variants with an abrupt transition from a low-grade area to a high-grade sarcoma. This latter subtype is inherently associated with worse prognoses.
The identification of intrachromosomal NAB2–STAT6 gene fusion as a pathognomonic molecular feature of SFT in 2013 [8,9,10] contributed to unequivocally unify under the unique term SFT other supposed distinct conditions, such as hemangiopericytoma and giant cell angiofibroma. In fact, for the purpose of definitive diagnostic confirmation, nuclear staining for STAT6 or the demonstration of NAB2–STAT6 gene fusion is required. As SFT can mimic many mesenchymal (and non-mesenchymal) tumors, the availability of these tools has become crucial for an accurate diagnosis of SFT [11]. The genomic fusions between NGFI-A-binding protein 2 (NAB2) and signal transducer and activator of transcription 6 (STAT6) can involve several exons in both genes, but they always produce a protein transcript without at least one repressor domain from NAB2, which is replaced by an activation domain of STAT6. Most investigators have attributed to NAB2 gene dysfunction the key genomic driver of the fusion transcript NAB2–STAT6. The lack of transcriptional corepression exerted on EGR1 by a NAB2 truncated gene would explain the increase in EGR1-mediated transcription in SFT [8,9].
In the report presented here, we describe a tumor lesion that exhibits histological features of SFT, exhibits nuclear positivity for STAT6 and carries a gene fusion, affecting STAT6 and the NFIX gene, another partner gene that is different than NAB2. The discovery of a new fusion partner in SFT opens intriguing new ways to refine the role of STAT6 in the sarcomatogenesis of this entity.
2. Results
2.1. Clinical and Pathological Features
A 74-year-old female, diagnosed in 2002 (at the age of 60 years old) with a deep-seated SFT in the pelvis, was sequenced for gene fusion detection with the Archer FusionPlexTM Sarcoma Panel, as a pathological central review procedure (at dept. Pathology, Hospital Virgen del Rocío) for enrollment in the GEIS-32 clinical trial (ClinicalTrials.gov: NCT02066285; EudraCT number: 2013-005456-15).
The patient was diagnosed in 2002 and treated with primary radical surgery. In 2009, she presented a local relapse and was treated with surgery, which included a bladder partial resection as well as a hysterectomy and bilateral oophorectomy. In 2010, she developed a new pelvic relapse, which was treated with an R1 resection. In subsequent scans, new evidence of disease was seen progressing slowly until June 2014, when a debulking surgery was performed due to symptomatic progression.
The patient was enrolled in the GEIS 32 clinical trial in May 2016 and started therapy with pazopanib 800 mg/day, achieving stable disease by CHOI and RECIST 1.1 evaluations, but treatment was interrupted due to grade 3 hepatic toxicity. Progression-free survival was 3.37 months. Post-protocol treatments included sunitinib, from October 2016 to May 2017, which achieved a partial response that led to a new surgery performed in June 2017. The patient was followed up without evidence of disease until January 2019, with slow progression in subsequent CT scans. In December 2019, a new surgical procedure was performed and since then, the patient has been under close surveillance for slowly progressing disease located in the pelvis. At the time of the last follow up in January 2021, the patient was alive and asymptomatic.
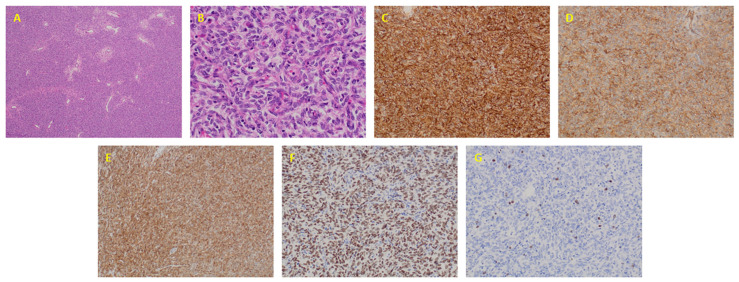
The local pathology report of the 2014 excisional biopsy described a hypercellular spindle cell neoplasm with hemangiopericytoid vascular pattern, with monotonous spindle to ovoid cells, without nuclear pleomorphism or tumor necrosis and with a low proliferative index. Complementary studies showed diffuse immunostaining for CD34, heterogeneous expression for CD99, diffuse expression of bcl-2 and diffuse and strong nuclear expression of STAT6; proliferative rate (Ki67) less than 2% (Figure 1).
Figure 1.
Pathological characterization of solitary fibrous tumor case. (A) Hypercellular spindle cell neoplasm with hemangiopericytoid vascular pattern (4×); (B) Monotonous spindle to ovoid cells, without nuclear pleomorphism (40×); (C) Diffuse expression of CD34 (20×); (D) Heterogeneous expression of CD99 (20×); (E) Diffuse expression of bcl-2 (20×); (F) Diffuse and strong nuclear expression of STAT6 (20×) and (G) Ki-67 proliferative index (20×).
2.2. Fusion Description
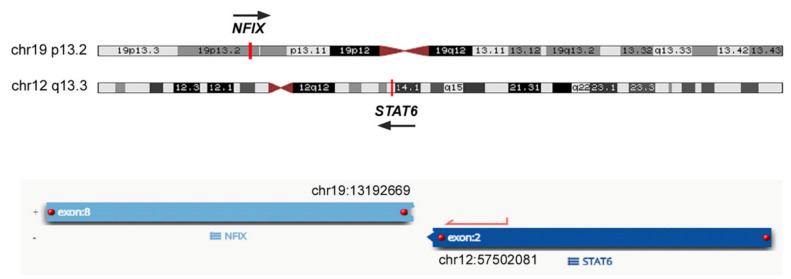
The tumor excised in 2014 was sequenced for gene fusion and breakpoint variant detection with the Archer FusionPlexTM Sarcoma Panel (Boulder, CO, USA). The results from FFPE tumor sample sequencing reveal a novel fusion transcript involving Nuclear Factor I X (NFIX), mapping to 19p13.2 and STAT6, mapping to 12q13.3. Total sample reads were 1,140,402. The bioinformatics analysis detected 378 unique reads spanning the NFIXex8–STAT6ex2 breakpoint with 55 different start sites, representing 59.5% of the reads intersecting the genomic location on either side of the breakpoint (Figure 2). This new fusion gene was detected in a non-treated surgical specimen (after the fourth surgery). No other gene fusion transcripts were detected in the same sample.
Figure 2.
Genomic location on either side of the breakpoint of the NFIXex8–STAT6ex2 gene fusion.
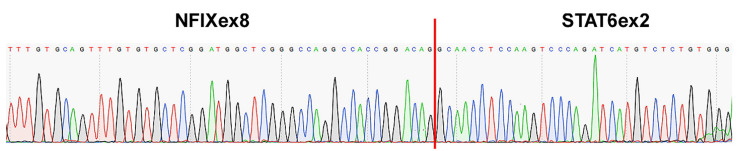
The new NFIX–STAT6 gene fusion and its specific breakpoint were validated using a set of primers for RT-PCR. Sanger sequencing was performed on the amplicon product and the sequence obtained was identical to the one founded by next generation sequencing (NGS). The breakpoint involved the exon 8 of NFIX and the exon 2 of STAT6 (Figure 3).
Figure 3.
Sanger sequencing validation of NFIXex8–STAT6ex2 gene fusion.
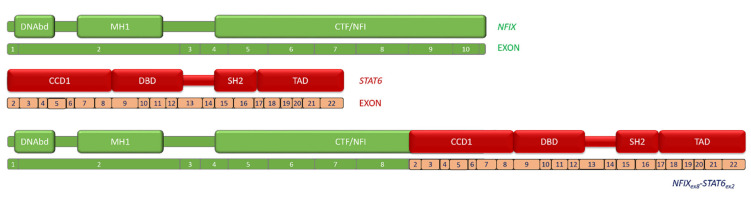
The in-frame breakpoints of the two genes occurred at nucleotide 1332 for NFIX (NM_001271043.2) and nucleotide 254 for STAT6 (NM_001178078.2). The novel fusion transcript maintained both the MAD homology 1 (MH1) domain and the N-terminal DNA-binding (DNAbd) domain in the N-terminus of NFIX and a larger part of its CAAT-box transcription factor–nuclear factor I (CTF-NFI) domain. The complete STAT6 protein is present in the NFIX–STAT6 fusion (Figure 4).
Figure 4.
Schematic representation of NFIX–STAT6 fusion. DNAbd: N-terminal DNA-binding domain; MH1: MAD homology 1 domain; CTF/ NFI: CAAT-box transcription factor–nuclear factor I domain; CCD1: Coiled-coil domain 1; DBD: DNA-binding domain; SH2: Src homology 2; and TAD: transcriptional activator domain.
3. Discussion
In this report, a novel fusion gene, NFIX–STAT6, is described in a patient diagnosed with a tumor that histopathologically and by immunohistochemistry exhibits characteristic traits of SFT. To the best of our knowledge, this is the first time ever that this fusion has been detected in cancer or any other disease, and is the first time that a different partner for STAT6, apart from NAB2, has been identified in SFT. This new fusion gene would suggest that STAT6 is also relevant in the potential mechanisms of SFT oncogenesis and progression, independently of the potential pivotal role of NAB2 as the driver of the tumorigenic processes of SFT within the NAB2–STAT6 fusion gene [9,10].
The majority of these fusions replace the chromodomain helicase DNA-binding protein 4 (CHD4) interacting domain (CID) in the C-terminus of NAB2, but they can also affect the N-terminus, replacing the NAB2 conserved domain 2 (NCD2). These two domains (i.e., CID and NCD2) are important for transcriptional repression and as a consequence of their lack of activity, and since early growth response 1 (EGR1)-interacting NCD1 is always present in the fusion protein, the EGR signaling pathway is normally positively dysregulated in SFT [9,10]. However, even if the key oncogenic role of NAB2–STAT6 may be related to the lack of repressive activity of NAB2 upon its targets, STAT6 dysfunction could also play an important part in SFT development. In fact, STAT6 seems to play a significant role in transcription and inflammation [12,13], processes linked to cancer development and progression [14,15,16,17,18].
According to NAB2–STAT6 fusion breakpoints, several different variants have been described; the two most frequent variants are the NAB2exon4–STAT6 exon2 (NAB2ex4–STAT6ex2) and the NAB2exon6–STAT6exon16/17 (NAB2ex6–STAT6ex16/17) [19]. The NAB2ex4–STAT6ex2 fusion lacks almost entirely the CID domain of NAB2, while STAT6 is almost complete. The NAB2ex6–STAT6ex16/17 lacks only the CID-exon 7 of NAB2, whereas STAT6 only keeps the last 5/6 exons, which includes part of the Src homology 2 domain (SH2) and the transcriptional activator domain (TAD). The SFTs harboring the NAB2ex4–STAT6ex2 variant are typically located in the thoracic cavity, have significantly larger tumor diameter (median 10 cm), exhibit the appearance of predominantly fibrotic and paucicellular features and the patients are normally older compared to the patients with other breakpoint variants. On the other hand, SFTs harboring the NAB2ex6–STAT6ex16/17 variant are commonly founded in the pelvis, meninges or extremities, have small tumor diameter (median 4.3 cm), show higher tumor cellularity compared to the previous fusion breakpoint variant and the patients are younger than those with the NAB2ex4–STAT6ex2 fusion [19,20]. However, and despite the more frequent association of the NAB2ex4–STAT6ex2 variant with less aggressive SFT, there is no unequivocal significant prognostic correlation between fusion breakpoints and survival [21]. Curiously, the fusion gene variant with the hardly truncated NAB2 gene, the NAB2ex6–STAT6ex16/17 variant, is associated with SFT features of higher aggressiveness. Besides NAB2–STAT6, no other fusion genes had been described in SFT until now.
STAT6 is a member of the signal transducer and activator of transcription family of proteins that is activated, in response to IL-4 or IL-13, by phosphorylation of tyrosine residues. These post-translational modifications promote homodimerization or heterotrimerization with other transcription factors, which translocate into the nucleus to initiate transcription [22]. Even when it is also recognized that non-phosphorylated STAT6 can be transcriptionally active [23], the STAT6 nuclear trafficking is tightly regulated. STAT6 is a predominantly cytoplasmic protein in a variety of cell types, as well as in tumors other than SFT [24]. The nuclear relocation of STAT6, which NAB2 localizes to the nucleus, with the transcript NAB2–STAT6, could lead to the activation of target genes of STAT6.
The pathway (IL-4/ STAT6) is important, for example, for type 2 T helper (Th2) cells’ development that can induce the differentiation of macrophages to an M2 phenotype [25,26]. Of note, M2 polarized macrophages seem to have an important prognostic role in SFT in two recent publications taken from a prospective multicenter phase II clinical trial of pazopanib for the treatment of advanced SFT [27,28]. Patients with high expression of CD209 had worse prognoses in terms of PFS and overall survival (OS) in the typical SFT cohort, while ISG15 high expression was associated with worse survival in the malignant/dedifferentiated SFT cohort. CD209 encodes a transmembrane receptor on the surface of both dendritic cells and macrophages, and it was defined as a marker of M2 polarized macrophages [29] and ISG15, an interferon-stimulated gene with a role in protein degradation and stemness, which seems to be involved in cell migration and CD8+ T cell immunosuppression by inducing M2-like macrophages [30]. Accordingly, it is plausible that STAT6-dependent regulation of macrophage polarization towards an M2-like phenotype may play an important role in SFT tumorigenesis and tumor progression.
Of note, the NFIX–STAT6 gene fusion maintained the complete STAT6 protein (exon 2 to 22), supporting the relevance of this transcription factor in the oncogenic processes driven by the chimeric RNA.
The NFIX gene is a member of the NFI (nuclear factor 1) transcription factor family, along with three other related genes involved in the regulation of stem cell biology during development. Initially, NFIX was identified as a transcription factor for fetal myogenesis; however, it is now recognized for its involvement in neural development and hematopoiesis [31]. NFIX gene expression has been shown to be significantly lower in some tumors (medulloblastoma, colorectal cancer or glioma), where it has been implicated as a tumor suppressor gene [32]. We can speculate that the nuclear protein NFIX could have lost its repressor function in some mesenchymal stem cells in the transcript, and similarly to NAB2, the transcript would retain STAT6 in the nucleus.
Gene fusions involving members of the NFI family have been reported in different types of cancer. However, functional relevance remains to be determined for most of them [33]. A recent report on secretory carcinoma of the skin described a novel NFIX–PKN1 translocation, but again, its oncogenic properties are not known [34], thus reducing our knowledge regarding its potential oncogenicity.
Finally, as besides NAB2–STAT6, no other fusion genes have been described in SFT, the newly detected NFIX–STAT6 fusion opens the inquiry to concede more relevance than just a secondary role to the STAT6 gene. This implies the opportunity for a wider perspective of new potential targets, such as macrophage implication in SFT progression for instance. This new discovery also spotlights the NFIX gene for deeper investigation of plausible signaling pathways related to SFT pathogenesis.
4. Material and Methods
4.1. Targeted RNA-Seq Library Preparation and Sequencing
Total nucleic acid was extracted with an Agencourt FormaPure kit (A33341; Beckman Coulter, Indianapolis, IN, USA), according to the manufacturers’ protocol. The extracted RNA was quantified using the Qubit® RNA HS Assay kit in combination with a Qubit® fluorometer (Q32852; Thermo Fisher Scientific, Waltham, MA, USA). Targeted library was prepared using the Archer™ FusionPlex™ Sarcoma Panel (SK0082; ArcherDX, Boulder, CO, USA) based on a targeted enrichment method named anchored multiplex PCR (AMP). A total of 200 ng of RNA was used for library preparation. The Archer™ FusionPlex™ Sarcoma Panel simultaneously detects and identifies fusions of 26 genes associated with soft-tissue sarcoma, without prior knowledge of the fusion partners or the specific breakpoints. In summary, RNA was reverse transcribed using random primers, first strand cDNA was synthesized and RNA quality was assessed using the Archer PreSeq RNA QC assay. After second strand cDNA synthesis, end repair, A-tailing and adapter ligation, cDNA was amplified by two rounds of nested PCR using gene-specific primers. KAPA Library Quantification Kit (KK4824; KAPA Biosystems, Wilmington, MA, USA) was used to quantify the final libraries. These libraries were sequenced on an Illumina MiSeq with MiSeq 300v2 reagents (MS-102-2002; Illumina, San Diego, CA, USA) for paired-end reads, 150 base pair read and dual index reads. FASTQ files were analyzed using Archer analysis pipeline version 6.0.3.2 and genome assembly GRCh37/hg19 as reference.
4.2. RT-PCR and Sanger Sequencing
Primers flanking the breakpoint sequence were designed with Primer3Plus [35], according to the reads from RNA sequencing. First, 450 ng of RNA was retro-transcribed using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). PCR was performed with Q5® Hot Start High-Fidelity 2X Master Mix (New England Biolabs, MA, USA) and the specific primers. The PCR products were analyzed by gel electrophoresis and purified using QIAquick PCR Purification Kit (Qiagen, Germany). Direct Sanger sequencing of PCR products was performed using BigDye Terminator v3.1 chemistry. Primer forward: AGCAGTCGAGCCCGTATTTC and primer reverse: ACCAGACCCCACAGAGACAT (product length 163bp).
4.3. Immunohistochemistry
The resected specimen was fixed in 10% neutral buffered formalin. Paraffin sections were stained with hematoxylin and eosin for routine histology. Immunohistochemical studies were performed on paraffin-embedded tissue sections using an automated OMNIS immunostainer (Agilent Dako, CA, USA), followed by antibody detection using the Dako EnVision+System and 3,3′-diaminobenzidine as a chromogen. The primary antibodies, clone, dilutions and sources used in this study are listed in Table 1. Appropriate positive and negative tissue controls were used throughout.
Table 1.
Immunohistochemical reagents.
| Antigen | Clone | Dilution | Source |
|---|---|---|---|
| Bcl-2 | 124 | Ready to use | Agilent |
| CD34 | QBEnd 10 | Ready to use | Agilent |
| CD99 | 12E7 | Ready to use | Agilent |
| STAT6 | Polyclonal | 1:50 | Gennova |
Acknowledgments
D.S.M. is the recipient of a Sara Borrell postdoctoral fellowship funded by the National Institute of Health Carlos III (ISCIII) (CD20/00155). J.L.M.-H. is the recipient of a PFIS predoctoral fellowship funded by the National Institute of Health Carlos III (ISCIII) (FI19/00184). The authors would also like to thank SELNET project. SELNET has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No. 825806. The authors would like to thank the Instituto de Salud Carlos III (ISCIII)–Fondo Europeo de Desarrollo Regional (FEDER), project reference PI18/01728.
Author Contributions
Conceptualization, D.S.M. and J.M.-B.; Methodology, D.S.M., J.D.-M., S.B., R.O.-F., J.L.M.-H., C.S.-A., F.R., N.H., C.D.M.F. and J.M.-B.; Validation, all the authors; Formal Analysis, D.S.M., J.D.-M. and J.M.-B.; Investigation, all the authors; Resources, all the authors; Data Curation, all the authors; Writing—Original Draft Preparation, D.S.M., J.D.-M., R.O.-F., A.S. and J.M.-B.; Writing—Review and Editing, all the authors; Visualization, all the authors; Supervision, D.S.M. and J.M.-B.; Project Administration, J.M.-B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board.
Informed Consent Statement
Informed consent was obtained from the subject involved in this study.
Data Availability Statement
Not applicable.
Conflicts of Interest
D.S.M. reports institutional research grants from PharmaMar, Eisai, Immix BioPharma and Novartis outside the submitted work; travel support from PharmaMar, Eisai, Celgene, Bayer and Pfizer. F.R. reports speaker, consultancy or advisory role fees from Roche, BMS, MSD, Merck, Novartis, AstraZeneca, Pfizer, Lilly, Amgen, Pierre-Fabre, Incyte, Daichii-Sankyo, Bayer and Janssen. NH reports grants, personal fees and non-financial support from PharmaMar, research grants from Eisai, Immix BioPharma and Novartis outside the submitted work and re-search funding for clinical studies (institutional) from PharmaMar, Eli Lilly and Company, AROG, Bayer, Eisai, Lixte, Karyopharm, Deciphera, GSK, Novartis, Blueprint, Nektar, Forma, Amgen and Daichii-Sankyo. J.M.-B. reports research grants from PharmaMar, Eisai, Immix BioPharma and Novartis outside the submitted work; honoraria for advisory board participation and expert testimony from PharmaMar, honoraria for advisory board participation from Eli Lilly and Company, Bayer and Eisai; and research funding for clinical studies (institutional) from PharmaMar, Eli Lilly and Company, AROG, Bayer, Eisai, Lixte, Karyopharm, Deciphera, GSK, Novartis, Blueprint, Nektar, Forma, Amgen and Daichii-Sankyo.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Klemperer P., Coleman B.R. Primary neoplasms of the pleura. A report of five cases. Am. J. Ind. Med. 1992;22:1–31. doi: 10.1002/ajim.4700220103. [DOI] [PubMed] [Google Scholar]
- 2.Rao N., Colby T.V., Falconieri G., Cohen H., Moran C.A., Suster S. Intrapulmonary solitary fibrous tumors: Clinicopathologic and immunohistochemical study of 24 cases. Am. J. Surg. Pathol. 2013;37:155–166. doi: 10.1097/PAS.0b013e31826a92f5. [DOI] [PubMed] [Google Scholar]
- 3.WHO Classification of Tumours Editorial Board . WHO Classification of Tumours Series. 5th ed. Volume 3. International Agency for Research on Cancer; Lyon, France: 2020. [(accessed on 10 April 2021)]. Soft Tissue and Bone Tumours. Available online: https://publications.Iarc.fr/588. [Google Scholar]
- 4.Fletcher C., Bridge J.A., Hogendoorn P.C.W., Mertens F. WHO Classification of Tumours of Soft Tissue and Bone. Volume 5 International Agency for Research on Cancer; Lyon, France: 2013. [Google Scholar]
- 5.O’Neill A.C., Tirumani S.H., Do W., Keraliya A.R., Hornick J., Shinagare A.B., Ramaiya N.H. Metastatic Patterns of Solitary Fibrous Tumors: A Single-Institution Experience. Am. J. Roentgenol. 2017;208:2–9. doi: 10.2214/AJR.16.16662. [DOI] [PubMed] [Google Scholar]
- 6.Demicco E.G., Park M.S., Araujo D.M., Fox P.S., Bassett R.L., Pollock R., Lazar A., Wang W.-L. Solitary fibrous tumor: A clinicopathological study of 110 cases and proposed risk assessment model. Mod. Pathol. 2012;25:1298–1306. doi: 10.1038/modpathol.2012.83. [DOI] [PubMed] [Google Scholar]
- 7.Demicco E.G., Wagner M.J., Maki R.G., Gupta V., Iofin I., Lazar A.J., Wang W.L. Risk assessment in solitary fibrous tumors: Validation and refinement of a risk stratification model. Mod. Pathol. 2017;30:1433–1442. doi: 10.1038/modpathol.2017.54. [DOI] [PubMed] [Google Scholar]
- 8.Chmielecki J., Crago A., Rosenberg M., O’Connor R., Walker S.R., Ambrogio L., Auclair D., McKenna A., Heinrich M., Frank D.A., et al. Whole-exome sequencing identifies a recurrent NAB2-STAT6 fusion in solitary fibrous tumors. Nat. Genet. 2013;45:131–132. doi: 10.1038/ng.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson D.R., Wu Y.-M., Kalyana-Sundaram S., Cao X., Lonigro R.J., Sung Y.-S., Chen C.-L., Zhang L., Wang R., Su F., et al. Identification of recurrent NAB2-STAT6 gene fusions in solitary fibrous tumor by integrative sequencing. Nat. Genet. 2013;45:180–185. doi: 10.1038/ng.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohajeri A., Tayebwa J., Collin A., Nilsson J., Magnusson L., von Steyern F.V., Brosjö O., Domanski H.A., Larsson O., Sciot R., et al. Comprehensive genetic analysis identifies a pathognomonic NAB2/STAT6 fusion gene, nonrandom secondary genomic imbalances, and a characteristic gene expression profile in solitary fibrous tumor. Genes Chromosomes Cancer. 2013;52:873–886. doi: 10.1002/gcc.22083. [DOI] [PubMed] [Google Scholar]
- 11.Doyle L.A., Vivero M., Fletcher C.D.M., Mertens F., Hornick J. Nuclear expression of STAT6 distinguishes solitary fibrous tumor from histologic mimics. Mod. Pathol. 2014;27:390–395. doi: 10.1038/modpathol.2013.164. [DOI] [PubMed] [Google Scholar]
- 12.Li J., Rodriguez J.P., Niu F., Pu M., Wang J., Hung L.-W., Shao Q., Zhu Y., Ding W., Liu Y., et al. Structural basis for DNA recognition by STAT6. Proc. Natl. Acad. Sci. USA. 2016;113:13015–13020. doi: 10.1073/pnas.1611228113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delgado-Ramirez Y., Colly V., Gonzalez G.V., Leon-Cabrera S. Signal transducer and activator of transcription 6 as a target in colon cancer therapy (Review) Oncol. Lett. 2020;20:455–464. doi: 10.3892/ol.2020.11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grivennikov S.I., Greten F.R., Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vinay D.S., Ryan E.P., Pawelec G., Talib W.H., Stagg J., Elkord E., Lichtor T., Decker W.K., Whelan R.L., Kumara H.M.C.S., et al. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Semin. Cancer Biol. 2015;35:S185–S198. doi: 10.1016/j.semcancer.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez H., Hagerling C., Werb Z. Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes Dev. 2018;32:1267–1284. doi: 10.1101/gad.314617.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox P.M., Goding C.R. Transcription and cancer. Br. J. Cancer. 1991;63:651–662. doi: 10.1038/bjc.1991.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bushweller J.H. Targeting transcription factors in cancer—from undruggable to reality. Nat. Rev. Cancer. 2019;19:611–624. doi: 10.1038/s41568-019-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tai H.-C., Chuang I.-C., Chen T.-C., Li C.-F., Huang S.-C., Kao Y.-C., Lin P.-C., Tsai J.-W., Lan J., Yu S.-C., et al. NAB2-STAT6 fusion types account for clinicopathological variations in solitary fibrous tumors. Mod. Pathol. 2015;28:1324–1335. doi: 10.1038/modpathol.2015.90. [DOI] [PubMed] [Google Scholar]
- 20.Barthelmeß S., Geddert H., Boltze C., Moskalev E.A., Bieg M., Sirbu H., Brors B., Wiemann S., Hartmann A., Agaimy A., et al. Solitary fibrous tumors/hemangiopericytomas with different variants of the NAB2-STAT6 gene fusion are characterized by specific histomorphology and distinct clinicopathological features. Am. J. Pathol. 2014;184:1209–1218. doi: 10.1016/j.ajpath.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 21.Vogels R., Macagno N., Griewank K., Groenen P., Verdijk M., Fonville J., Kusters B., French CNS SFT/HPC Consortium. Dutch CNS SFT/HPC Consortium. Figarella-Branger D., et al. Prognostic significance of NAB2-STAT6 fusion variants and TERT promotor mutations in solitary fibrous tumors/hemangiopericytomas of the CNS: Not (yet) clear. Acta Neuropathol. 2019;137:679–682. doi: 10.1007/s00401-019-01968-3. [DOI] [PubMed] [Google Scholar]
- 22.Schmitz J., Radbruch A. Immunoglobulin class switching. In: Delves P.J., editor. Encyclopedia of Immunology. 2nd ed. Elsevier; Oxford, UK: 1998. pp. 1302–1306. [Google Scholar]
- 23.Sehgal P.B. Paradigm shifts in the cell biology of STAT signaling. Semin. Cell Dev. Biol. 2008;19:329–340. doi: 10.1016/j.semcdb.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schweizer L., Koelsche C., Sahm F., Piro R., Capper D., Reuss D.E., Pusch S., Habel A., Meyer J., Göck T., et al. Meningeal hemangiopericytoma and solitary fibrous tumors carry the NAB2-STAT6 fusion and can be diagnosed by nuclear expression of STAT6 protein. Acta Neuropathol. 2013;125:651–658. doi: 10.1007/s00401-013-1117-6. [DOI] [PubMed] [Google Scholar]
- 25.Yu T., Gan S., Zhu Q., Dai D., Li N., Wang H., Chen X., Hou D., Wang Y., Pan Q., et al. Modulation of M2 macrophage polarization by the crosstalk between Stat6 and Trim24. Nat. Commun. 2019;10:4353. doi: 10.1038/s41467-019-12384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elo L., Järvenpää H., Tuomela S., Raghav S., Ahlfors H., Laurila K., Gupta B., Lund R.J., Tahvanainen J., Hawkins R.D., et al. Genome-wide Profiling of Interleukin-4 and STAT6 Transcription Factor Regulation of Human Th2 Cell Programming. Immunity. 2010;32:852–862. doi: 10.1016/j.immuni.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Martin-Broto J., Cruz J., Penel N., Le Cesne A., Hindi N., Luna P., Moura D., Bernabeu D., de Alava E., Lopez-Guerrero J.A., et al. Pazopanib for treatment of typical solitary fibrous tumours: A multicentre, single-arm, phase 2 trial. Lancet Oncol. 2020;21:456–466. doi: 10.1016/S1470-2045(19)30826-5. [DOI] [PubMed] [Google Scholar]
- 28.Martin-Broto J., Stacchiotti S., Lopez-Pousa A., Redondo A., Bernabeu D., de Alava E., Casali P.G., Italiano A., Gutierrez A., Moura D., et al. Pazopanib for treatment of advanced malignant and dedifferentiated solitary fibrous tumour: A multicentre, single-arm, phase 2 trial. Lancet Oncol. 2019;20:134–144. doi: 10.1016/S1470-2045(18)30676-4. [DOI] [PubMed] [Google Scholar]
- 29.Rőszer T. Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediat. Inflamm. 2015;2015:816460. doi: 10.1155/2015/816460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen R.-H., Xiao Z.-W., Yan X.-Q., Han P., Liang F.-Y., Wang J.-Y., Yu S.-T., Zhang T.-Z., Chen S.-Q., Zhong Q., et al. Tumor cell-secreted ISG15 promotes tumor cell migration and immune suppression by inducing the macrophage M2-Like Phenotype. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.594775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piper M., Gronostajski R., Messina G. Nuclear factor one X in development and disease. Trends Cell Biol. 2019;29:20–30. doi: 10.1016/j.tcb.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Liu S., Qu D., Li W., He C., Li S., Wu G., Zhao Q., Shen L., Zhang J., Zheng J. miR647 and miR1914 promote cancer progression equivalently by downregulating nuclear factor IX in colorectal cancer. Mol. Med. Rep. 2017;16:8189–8199. doi: 10.3892/mmr.2017.7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen K.S., Lim J.W., Richards L.J., Bunt J. The convergent roles of the nuclear factor I transcription factors in development and cancer. Cancer Lett. 2017;410:124–138. doi: 10.1016/j.canlet.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 34.Kastnerova L., Luzar B., Goto K., Grishakov V., Gatalica Z., Kamarachev J., Martinek P., Hájková V., Grossmann P., Imai H., et al. Secretory carcinoma of the skin: Report of 6 Cases, including a case with a novel NFIX-PKN1 translocation. Am. J. Surg. Pathol. 2019;43:1092–1098. doi: 10.1097/PAS.0000000000001261. [DOI] [PubMed] [Google Scholar]
- 35.Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.