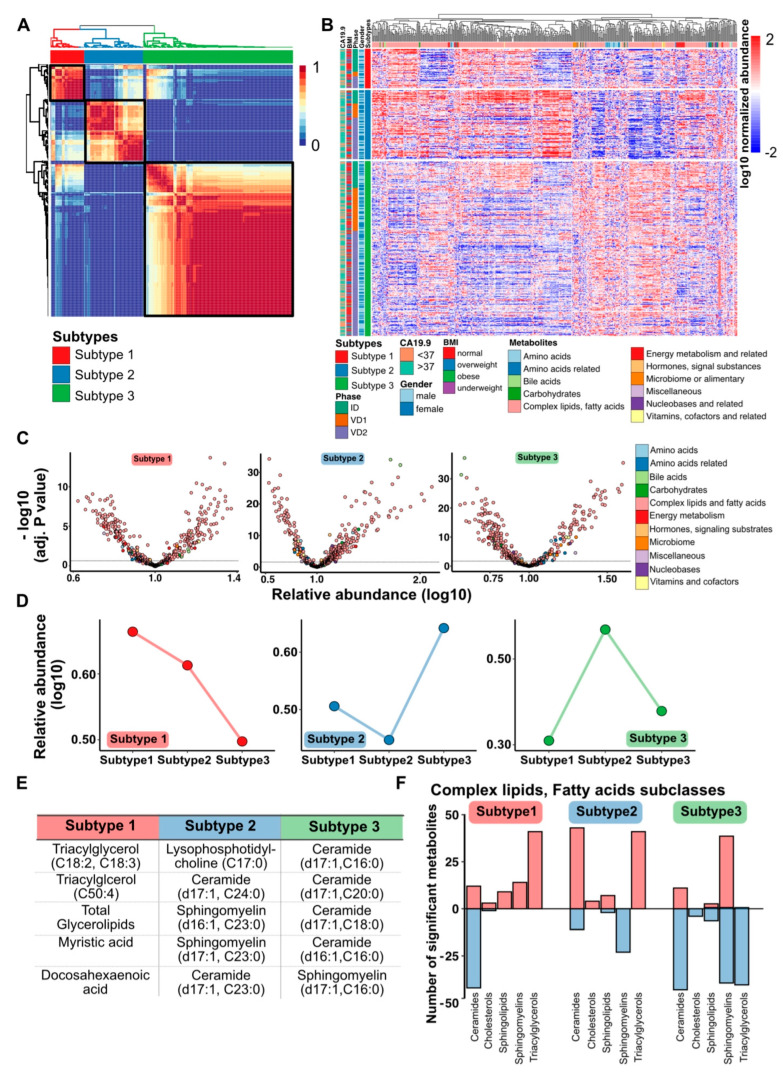
Figure 2.
Metabolic classification of the PDAC disease cohort: (A) unsupervised consensus classification of PDAC lipidomics data representing 361 patients using non-negative matrix factorization (NMF). (B) Heatmap of three consensus clusters based on plasma metabolome profiling. Data are shown as log10 normalized abundance of the metabolites. (C) Distribution of detected metabolites according to p-value in each subtype. Relative abundance is normalized by log10. (D) Median differential distributions of top 30 identified metabolites from each subtype. (E) Table shows a list of top 5 discriminative metabolites from each subtype. (F) Bar graph exhibits the 5 prominent subclasses from complex lipid and fatty acid classes, with significant fold changes per subtype compared to the rest. Red bar depicts upregulated metabolites, whereas blue bar denotes downregulated metabolites.