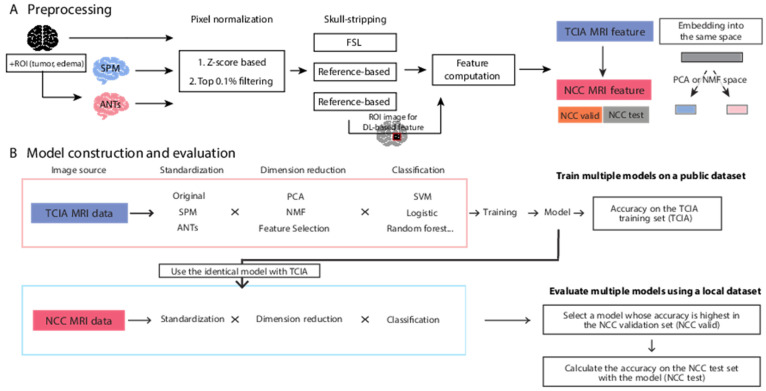
Figure 1.
An overview of the dataset preprocessing and analytical methods used in this study. (A) Brain image data is applied to the multiple preprocessing steps; standardization of brain shape, pixel normalization, and skull-stripping. Then image features for radiogenomics are computed. To test the robustness of the trained models, we separated the whole NCC dataset to two independent partial NCC dataset: NCC validation and test set. The embedding space for dimension reduction is obtained from the TCIA dataset and applied to the partial NCC datasets independently. (B) In a machine learning workflow, all ML models were generated from TCIA data. The models were applied to TCIA (for cross validation), with half of the NCC data used for validation (termed NCC validation or NCC valid) and the other half of the NCC data (NCC test).