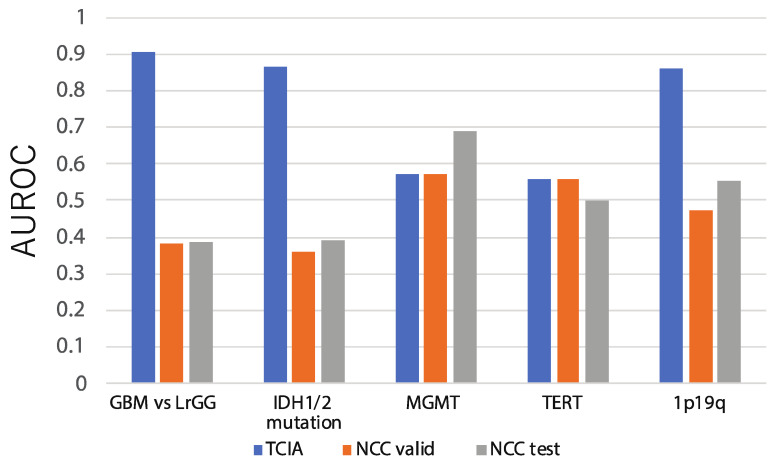
Figure 3.
AUROCs were used as criteria to assess prediction accuracies of our machine learning workflow, without pre-processing, for 5 prediction problems: GBM/LrGG classification, IDH mutation existence, MGMT methylation status prediction, TERT promoter methylation prediction, and chr 1p19q co-deletion prediction. The accuracy of cross-validation results from TCIA (depicted as TCIA in Figure 2), as well as the accuracy of the application of the model to the NCC validation set (NCC valid) and the NCC test set (NCC test), are indicated by blue, orange, and grey bars, respectively.