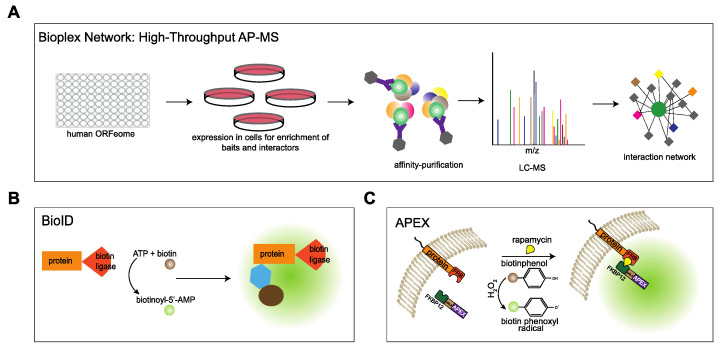
Figure 3.
Approaches used to analyze the interactome of the VAPs. (A) Schematic of high-throughput interaction mapping using affinity-purification mass spectrometry. From the human ORFeome, a lentiviral library was created. The baits were expressed in HEK293T cells, subjected to affinity purification, and protein complexes were analyzed by LC-MS. All the interactors and baits were combined to generate a network of the human interactome [43]. (B) In a BioID approach, the protein of interest is tagged with biotin ligase and expressed in cells. The enzyme converts free biotin into reactive biotinoyl-5′-AMP, which reacts with primary amines of proximal proteins. The biotinylated proteins are enriched and identified using mass spectrometry [59]. (C) In APEX-based proximity labelling, the protein of interest is fused to ascorbate peroxidase (APEX). In the presence of H2O2, APEX converts biotin phenol into biotin phenoxyl radicals, which covalently label proteins in close proximity. In RAPIDS, the specific subcellular interactome of VAPB was analyzed by using rapamycin induced dimerization of FKBP12- and FRB-containing fusion proteins, followed by APEX2-dependent biotinylation of proteins and their identification by MS [60]. In this approach, SILAC allows a direct comparison between plus- and minus-rapamycin conditions and, thus, a very efficient elimination of the background.