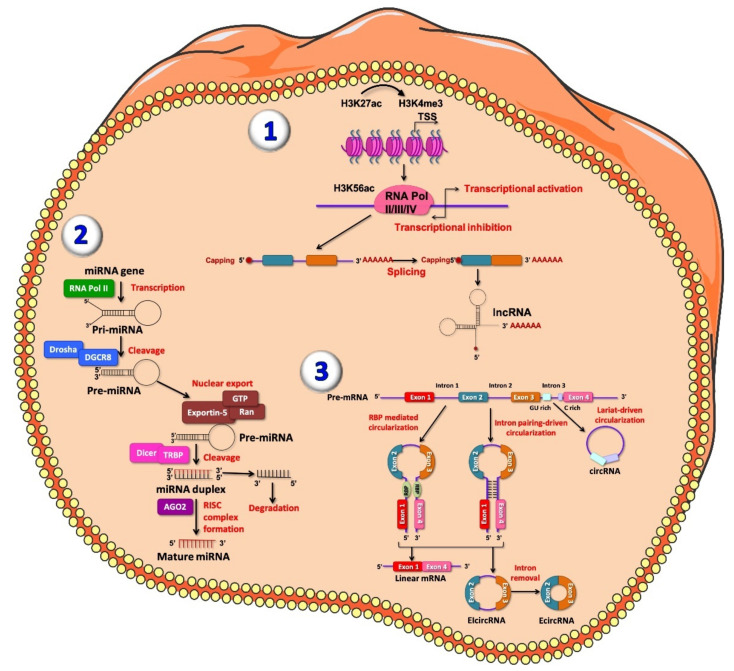
Figure 1.
Representative figure of long non-coding RNAs (lncRNAs) (1), microRNAs (miRNAs) (2) and circular RNAs (circRNA) (3) biogenesis. (1) At the chromatin state, H3K27ac and H3K4me3 are enriched at lncRNA promoter; transcription of lncRNA is initiated from different promoters in antisense direction, enriched for H3K56ac and Pol II/III/IV. The resulting pre-mature lncRNA is subjected to a 3′-polyadenylated and the 5′-end capping with methyl-guanosine. Then, all introns are spliced, resulting in a final mature lncRNA. (2) MiRNAs are firstly transcribed by RNA polymerase II into the nucleus, producing primary miRNAs (pri-miRNAs), a stem loop shaped RNA sequence. Pri-miRNA, once processed, is recognized and cleaved by the multi-protein complex Microprocessor within the nucleus. This complex is composed by two double-stranded molecules: RNase III enzyme DROSHA and RNA-binding protein DGCR8. DROSHA cuts, by its RNase III domains, in two different points of the double strand RNA (dsRNA) towards the base of the stem-loop generating a ~70 nucleotide hairpin–shaped precursor miRNAs (pre-miRNAs), showing an overhang at the 3′ end of 2 nucleotide left by the asymmetrical cut made by DROSHA recognized by Exportin-5 which carries the pre-miRNA into the cytoplasm. Here, the pre-miRNA is further processed by DICER/TRBP complex, which generates imperfect duplexes of 22 nucleotides containing a guide strand and a passenger strand. The guide strand (represented in red) together with Argonaute proteins forms RNA-induced silencing complex (RISC) and generates the mature miRNA, while the passenger strand is finally degraded. (3) CircRNAs are generated by an alternative splicing mechanism of pre-mRNA, termed back-splicing. In this process, the 3′-end of an exon binds to the 5′-end of its own or to an upstream exon through a 3′,5′- phosphodiester bond, forming a closed structure with a back-splicing junction site. Two models of circRNAs biogenesis have been described: the lariat model and the direct back-splicing model, further subdivided into RBP-mediated circularization and Intron pairing-driven circularization, regulating adjacent splice sites [52]. Lariat-driven circularization occurs through the interaction between the 3′ hydroxyl of the upstream exon with the 5′ phosphate of the downstream exon generating a covalent binding, producing a lariat containing both exons and introns. From both RBP-mediated circularization and intron pairing-driven circularization four main subtypes of circRNAs have been identified: exonic-circRNAs (ecircRNAs), mainly derived from single or multiple exons and exonic-intronic circRNAs (EIciRNAs), which consist of both introns and exons.