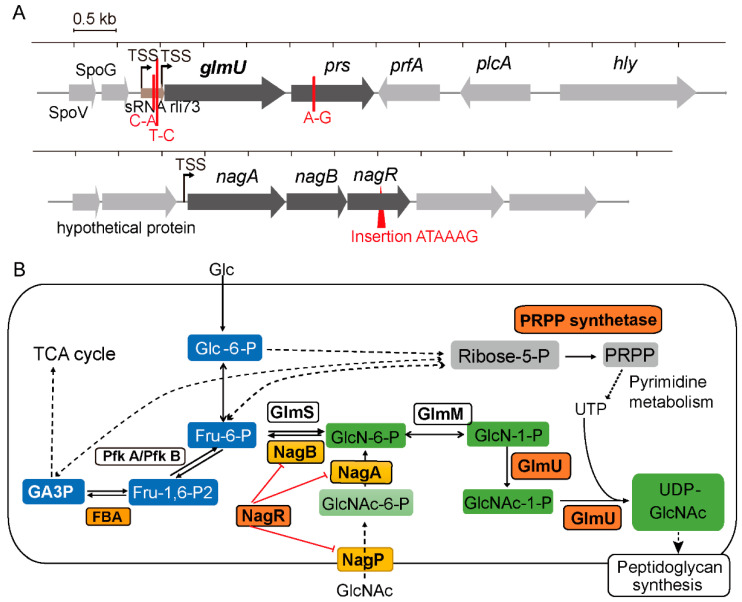
Figure 6.
(A) Locations of yvcK suppression mutations in the glmU-prs and nagABR operon. Single nucleotide variations are depicted as red bars and the in-frame insertion in nagR as a red triangle. Transcription start sites (TSSs) [25] of operons are indicated with a black arrow. (B) Scheme of the UDP-GlcNAc biosynthesis pathway with indication of the functions of GlmU and NagR [15,28]. Proteins affected by suppression mutations identified in this work are colored in orange. NagR-suppressed proteins are colored yellow. Intermediates of the glycolysis, the UDP-GlcNAc pathway and the pentose phosphate pathway are shown in blue, green and grey, respectively. PRPP, phospho-alpha-D-ribosyl-1-pyrophosphate; PRPP synthetase, ribose-phosphate pyrophosphokinase; Pfk, phosphofructokinase; and FBA, fructose-bisphosphate aldolase.