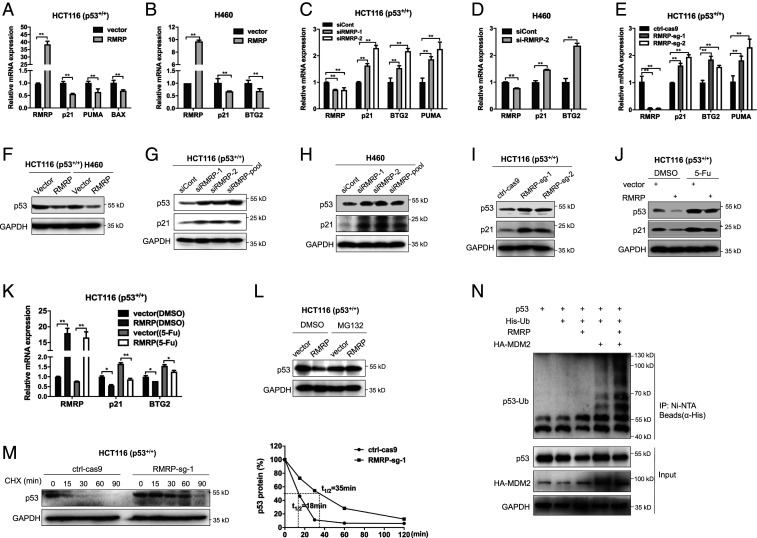
Fig. 2.
RMRP represses p53 activity by enhancing MDM2-induced p53 proteasomal degradation. (A and B) Overexpression of RMRP reduces p53 target gene expression in HCT116 p53+/+ and H460 cells. (C and D) Knockdown of RMRP increases p53 target gene expression in HCT116 p53+/+ and H460 cells. (E) CRISPR-Cas9–mediated ablation of RMRP induces p53 target gene expression in HCT116 p53+/+ cells. (F) Overexpression of RMRP decreases the protein level of p53 in HCT116 p53+/+ and H460 cells. (G and H) Knockdown of RMRP elevates the protein levels of p53 and p21 in HCT116 p53+/+ and H460 cells. (I) Knockout of RMRP induces p53 and p21 protein levels in HCT116 p53+/+ cell. (J and K) Overexpression of RMRP impairs 5-FU–induced p53 activation determined by IB (J) and RT-qPCR (K). (L) The proteasome inhibitor MG132 blocks RMRP-mediated p53 degradation in HCT116 p53+/+ cells. Cells were treated with MG132 (20 μM) for 6 h before harvested for IB. (M) The p53’s half-life is extended upon RMRP depletion. The ctrl-Cas9 and RMRP-sg-1 cell lines were treated with 100 μg/mL of cycloheximide (CHX) and harvested at the indicated time points for IB (Left). (Right) The ratios of p53/GAPDH. (N) RMRP promotes MDM2-dependent ubiquitination of p53. HCT116 p53−/− cells were transfected with combinations of plasmids encoding p53, RMRP, HA-MDM2, and His-Ub as indicated and treated with MG132 (20 μM) for 6 h before harvested for in vivo ubiquitination assay. *P < 0.05, **P < 0.01 by two-tailed Student’s t test.