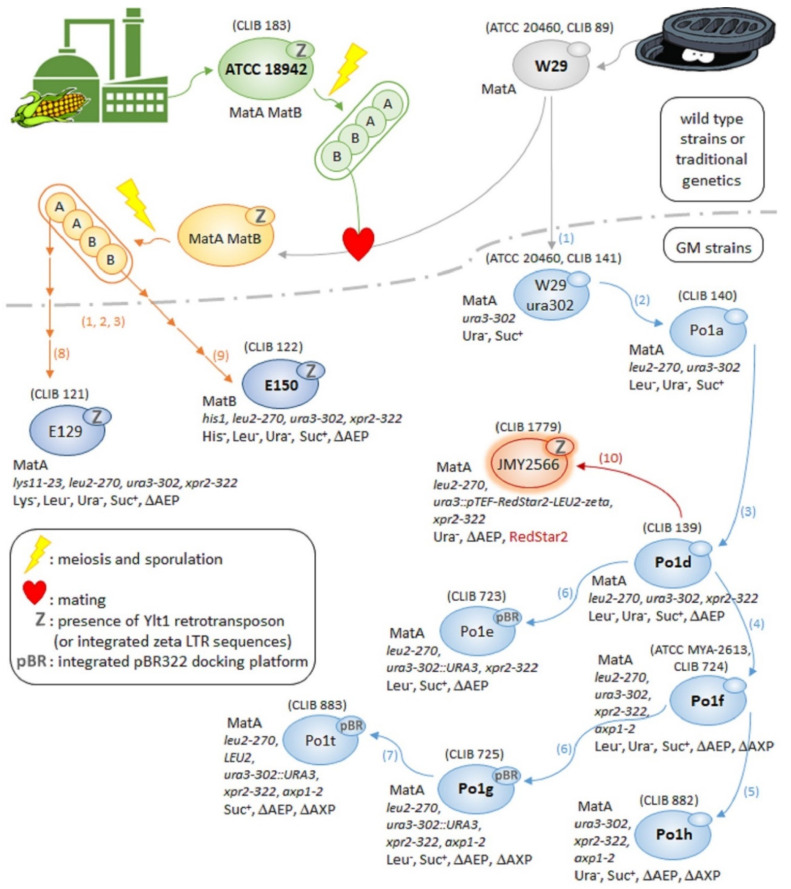
Figure 2.
Genealogy of strains of biotechnological interest derived from ATCC 18942 and/or W29. Schematic representation of the origins of ATCC 18942 and W29 wild-type strains and their publicly available derivatives: genotypes and phenotypes are indicated near each strain; yeast collection reference numbers are shown only for ATCC and CLIB-Levures (others can be found in Table 1). Genetic events are represented by the lightning and heart symbols; the presence of sequences that can be used for targeting integrations (zeta sequences, plasmidic backbone) is indicated in the nascent yeast bud. The genetic engineering steps are indicated by bracketed numbers, as follows: (1) genic conversion of URA3 into ura3-302 (corresponding to ura3::pXPR2:SUC2, namely to a 728 bp XhoI-EcoRV deletion in URA3 coding region with disruption by the SUC2 gene from S. cerevisiae under the control of the XPR2 promoter, conferring the ability to grow on sucrose or molasses) [15]; (2) genic conversion of LEU2 into leu2-270 (681 bp StuI deletion in LEU2 coding region); (3) genic conversion of XPR2 into xpr2-322 (149 bp ApaI deletion in XPR2 coding region, inactivating alkaline extracellular protease); (4) genic conversion of AXP1 into axp1-2 (655 bp NcoI deletion in AXP1 coding region, inactivating acid extracellular protease); (5) genic conversion of leu2-270 into LEU2 (restoration of wild-type allele); (6) integration of NcoI-linearized pINA300’ plasmid (URA3 gene in pBR322 [173]); (7) integration of NotI-linearized LEU2-carrying pINA1269 plasmid [159] at pBR docking platform; (8) genic conversion of LYS11 into lys11-23; (9) genic conversion of HIS1 into his1; (10) genic conversion of ura3-302 into ura3::pTEF-RedStar2-LEU2-zeta (replacement of the SUC2 expression cassette disrupting URA3 with two cassettes, for expressing the RedStar2 fluorescent protein from TEF promoter and for LEU2, and a zeta LTR sequences docking platform) [162]. The names of the more prominent strains are in bold type. Information gathered from [10,43] or ATCC and CLIB-Levures websites.