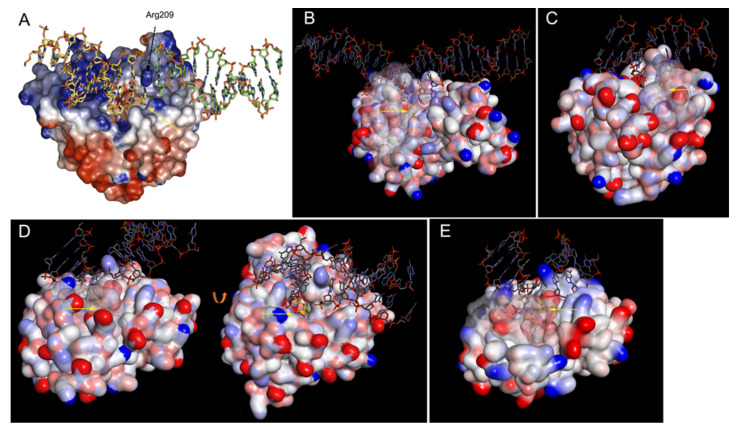
Figure 4.
Nucleotide flipping out of DNA helix is the conserved DNA base damage recognition mechanism among NIR AP endonucleases. (A) Enzyme–substrate complex of the Mth212 D151N mutant obtained with dsDNA (exonuclease reaction substrate with a terminal dC in the active site) (PDB: 3GA6) [162]. (B) PfuEndoQ complex with a 27 mer duplex substrate with dU at the active site (PDB: 7K30) [167]. (C) Human APE1 bound to non-cleaved THF-DNA (PDB: 1DE8) [141]. (D) E. coli Nfo H69A mutant bound to a cleaved DNA duplex containing αdA (PDB: 4K1G) [168]. (E) E. coli Nfo Y72A mutant bound to THF-DNA (PDB: 2NQ9) [169]. Yellow arrows point to the nucleotides flipped into the active sites.