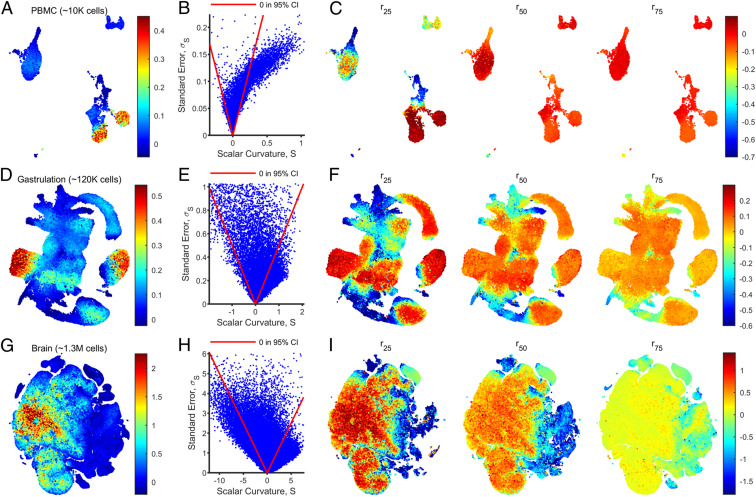
Fig. 4.
scRNAseq datasets have localized regions of nonzero scalar curvature. (A) Scalar curvatures were computed for a scRNAseq dataset with PBMCs collected from a healthy human donor. The ambient () and manifold () dimensions were specified to be eight and three, respectively, and variable neighborhood sizes were chosen by setting (SI Appendix, Supporting Methods, section F.6). The scalar curvatures are shown here overlaid onto UMAP coordinates, after smoothing the values over nearest neighbors in the ambient space. (B) Scatter plot of (unsmoothed) scalar curvatures, , and associated SEs, , for each data point in the PBMC dataset. Points enclosed by the red lines reported a 95% CI () including zero. (C) As in A, but with scalar curvatures computed by using a fixed neighborhood size, , for all data points. The value of was set to be the 25th, 50th, and 75th percentile values (left to right) of the neighborhood sizes used in A (SI Appendix, Fig. S4E). Points for which a neighborhood of size does not include enough neighbors for regression are not shown. (D–F) As in A–C for a mouse gastrulation dataset with , , and . (G–I) As in A–C for a mouse brain dataset with , , and , plotted on t-SNE coordinates.