Table 1.
Calculated binding affinities (in kcal/mol), 2D chemical structures and binding features for darunavir and the top five most promising cembranoid diterpenes against SARS-CoV-2 main protease (Mpro) a.
| Compound Name (Number) | Genus | 2D Chemical Structure | Docking Score (kcal/mol) | Binding Features b |
|---|---|---|---|---|
| Darunavir | --- |
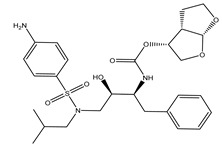
|
−8.2 | LEU167 (1.96 Å), GLU166 (1.94, 2.88 Å) |
| Sarelengan B (363) | S. elegans |
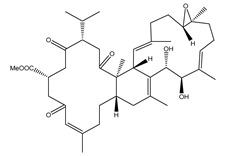
|
−9.8 | GLY143 (2.39 Å), GLU166 (1.94 Å), GLN189 (2.58 Å), THR190 (2.33 Å) |
| Bislatumlide A (340) | S. latum |
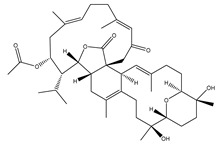
|
−9.6 | GLY143 (1.88 Å), GLU166 (2.68 Å) |
| Dioxanyalolide (347) | S. elegans |
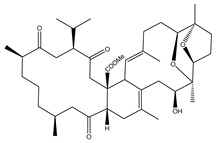
|
−9.5 | GLU166 (2.07 Å) |
| Desacetylnyalolide (345) | S. elegans |
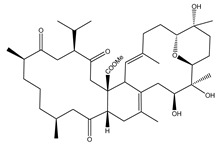
|
−9.1 | GLU166 (1.66, 2.12 Å), THR190 (2.42 Å) |
| Lobophytone W (357) | S. elegans |
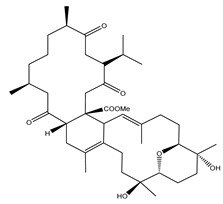
|
−8.7 | HIS41 (2.01 Å), CYS145 (2.34 Å), GLU166 (2.35, 2.86 Å) |
a The potent Sarcophyton cembranoid diterpenes metabolites were selected based on latter MM-GBSA binding energy calculations. b Only hydrogen bonds (in Å) were displayed.
